
DeBrazza s monkey arterivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Arnidovirineae; Arteriviridae; Simarterivirinae; Iotaarterivirus; Debiartevirus; Iotaarterivirus debrazmo
Average proteome isoelectric point is 7.74
Get precalculated fractions of proteins
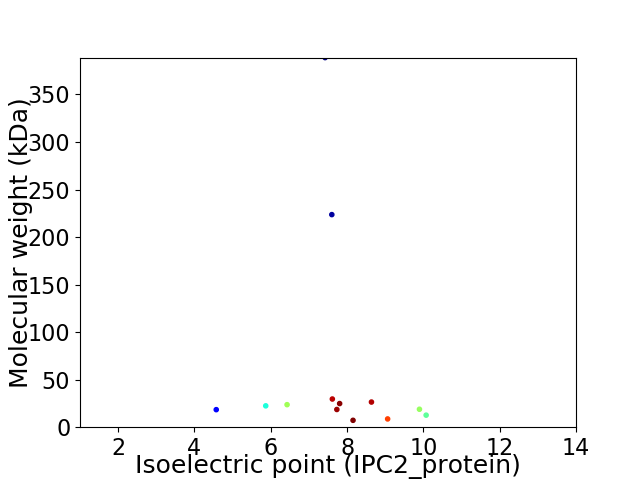
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B6CFJ6|A0A0B6CFJ6_9NIDO Nucleocapsid protein OS=DeBrazza's monkey arterivirus OX=1965063 GN=ORF7 PE=3 SV=1
MM1 pKa = 7.89GDD3 pKa = 3.14QLLRR7 pKa = 11.84CVTPSCGIFLLLLCVLSCASNQTYY31 pKa = 10.09ICLPSFNSSLLLTINASTHH50 pKa = 5.32SCLFTGNVLVGSQYY64 pKa = 11.5GLPICPQANEE74 pKa = 4.56RR75 pKa = 11.84YY76 pKa = 9.35FDD78 pKa = 4.14ANASGQLEE86 pKa = 4.98LPTGLDD92 pKa = 3.69LVFLRR97 pKa = 11.84YY98 pKa = 8.87VWQHH102 pKa = 6.1LDD104 pKa = 3.3QFPSLFNGSKK114 pKa = 10.38PEE116 pKa = 3.94QQASCWVINSTQPTSTPSLPQEE138 pKa = 3.9YY139 pKa = 10.72LEE141 pKa = 4.12FWEE144 pKa = 4.7FQPLPLLLLILVLQLVMLLPPLL166 pKa = 3.87
MM1 pKa = 7.89GDD3 pKa = 3.14QLLRR7 pKa = 11.84CVTPSCGIFLLLLCVLSCASNQTYY31 pKa = 10.09ICLPSFNSSLLLTINASTHH50 pKa = 5.32SCLFTGNVLVGSQYY64 pKa = 11.5GLPICPQANEE74 pKa = 4.56RR75 pKa = 11.84YY76 pKa = 9.35FDD78 pKa = 4.14ANASGQLEE86 pKa = 4.98LPTGLDD92 pKa = 3.69LVFLRR97 pKa = 11.84YY98 pKa = 8.87VWQHH102 pKa = 6.1LDD104 pKa = 3.3QFPSLFNGSKK114 pKa = 10.38PEE116 pKa = 3.94QQASCWVINSTQPTSTPSLPQEE138 pKa = 3.9YY139 pKa = 10.72LEE141 pKa = 4.12FWEE144 pKa = 4.7FQPLPLLLLILVLQLVMLLPPLL166 pKa = 3.87
Molecular weight: 18.45 kDa
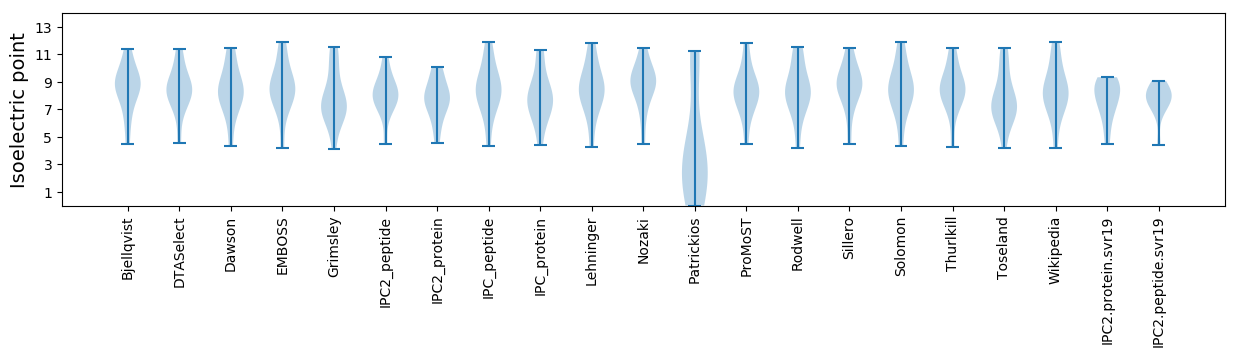
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B6CFI5|A0A0B6CFI5_9NIDO ORF2a' protein OS=DeBrazza's monkey arterivirus OX=1965063 GN=ORF2a' PE=4 SV=1
MM1 pKa = 7.87PSQTGARR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84VDD12 pKa = 3.61MVTTICSDD20 pKa = 3.29PGYY23 pKa = 6.71TTLAFTIAPVLLACLRR39 pKa = 11.84LFRR42 pKa = 11.84PSVRR46 pKa = 11.84GVVCLLCIATLAYY59 pKa = 10.23AATAFNEE66 pKa = 4.17HH67 pKa = 6.34SLATILTISFILVYY81 pKa = 10.65LSYY84 pKa = 9.74KK85 pKa = 7.7TVAWVVIRR93 pKa = 11.84CRR95 pKa = 11.84MCRR98 pKa = 11.84LGRR101 pKa = 11.84QYY103 pKa = 9.16ITAPSSMVEE112 pKa = 3.97TSLGRR117 pKa = 11.84SAITAGQSAVVRR129 pKa = 11.84RR130 pKa = 11.84TSGLTSANGLLVPDD144 pKa = 3.98VKK146 pKa = 10.77RR147 pKa = 11.84IILHH151 pKa = 5.61GRR153 pKa = 11.84VAAKK157 pKa = 10.18KK158 pKa = 10.45GLVNLRR164 pKa = 11.84KK165 pKa = 10.32YY166 pKa = 10.37GWQTKK171 pKa = 9.44NKK173 pKa = 9.82
MM1 pKa = 7.87PSQTGARR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84VDD12 pKa = 3.61MVTTICSDD20 pKa = 3.29PGYY23 pKa = 6.71TTLAFTIAPVLLACLRR39 pKa = 11.84LFRR42 pKa = 11.84PSVRR46 pKa = 11.84GVVCLLCIATLAYY59 pKa = 10.23AATAFNEE66 pKa = 4.17HH67 pKa = 6.34SLATILTISFILVYY81 pKa = 10.65LSYY84 pKa = 9.74KK85 pKa = 7.7TVAWVVIRR93 pKa = 11.84CRR95 pKa = 11.84MCRR98 pKa = 11.84LGRR101 pKa = 11.84QYY103 pKa = 9.16ITAPSSMVEE112 pKa = 3.97TSLGRR117 pKa = 11.84SAITAGQSAVVRR129 pKa = 11.84RR130 pKa = 11.84TSGLTSANGLLVPDD144 pKa = 3.98VKK146 pKa = 10.77RR147 pKa = 11.84IILHH151 pKa = 5.61GRR153 pKa = 11.84VAAKK157 pKa = 10.18KK158 pKa = 10.45GLVNLRR164 pKa = 11.84KK165 pKa = 10.32YY166 pKa = 10.37GWQTKK171 pKa = 9.44NKK173 pKa = 9.82
Molecular weight: 18.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7572 |
59 |
3587 |
582.5 |
63.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.509 ± 0.616 | 3.196 ± 0.204 |
4.226 ± 0.562 | 3.209 ± 0.203 |
4.437 ± 0.371 | 6.762 ± 0.719 |
2.773 ± 0.684 | 4.543 ± 0.375 |
3.711 ± 0.379 | 11.067 ± 0.6 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.373 ± 0.108 | 3.288 ± 0.362 |
6.194 ± 0.493 | 3.117 ± 0.367 |
5.137 ± 0.286 | 7.937 ± 0.578 |
6.894 ± 0.232 | 8.135 ± 0.625 |
1.149 ± 0.143 | 3.341 ± 0.222 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
