
Gracilibacillus orientalis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Gracilibacillus
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
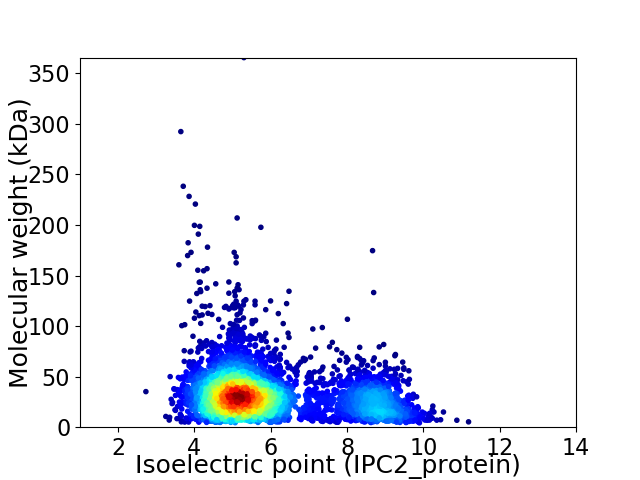
Virtual 2D-PAGE plot for 4278 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
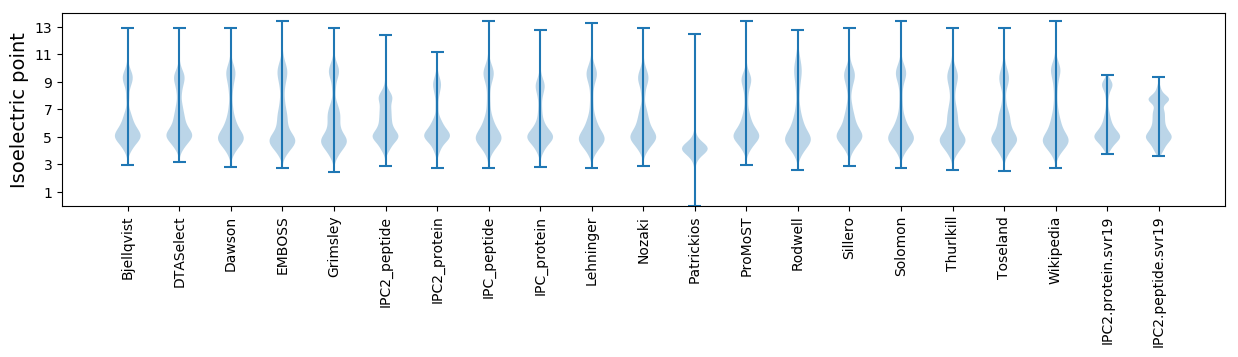
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4LTD3|A0A1I4LTD3_9BACI Drug resistance transporter EmrB/QacA subfamily OS=Gracilibacillus orientalis OX=334253 GN=SAMN04487943_105200 PE=3 SV=1
MM1 pKa = 7.6KK2 pKa = 10.25KK3 pKa = 9.68ILLIAILFTMSVVIMACSSDD23 pKa = 3.46EE24 pKa = 4.06ADD26 pKa = 3.33SVEE29 pKa = 4.39EE30 pKa = 3.93NVEE33 pKa = 4.05SDD35 pKa = 4.28GEE37 pKa = 4.36TEE39 pKa = 4.04EE40 pKa = 5.55QSDD43 pKa = 3.85GDD45 pKa = 4.13EE46 pKa = 3.92NTLYY50 pKa = 10.93DD51 pKa = 4.09EE52 pKa = 5.32SNLLSLGQSGSIEE65 pKa = 4.23ATNSSYY71 pKa = 11.2EE72 pKa = 3.93VTVEE76 pKa = 3.98SFEE79 pKa = 4.12VLNDD83 pKa = 3.24IVGQQSSEE91 pKa = 3.81EE92 pKa = 4.25VYY94 pKa = 10.7ILLDD98 pKa = 3.61YY99 pKa = 11.06QITNTGEE106 pKa = 3.69EE107 pKa = 4.61AIQGLAVTEE116 pKa = 4.33GNLFNAAGAKK126 pKa = 9.17GLSVTSYY133 pKa = 11.4DD134 pKa = 4.43FISIPDD140 pKa = 3.83TEE142 pKa = 4.39IKK144 pKa = 10.28PGEE147 pKa = 4.15SAAAQILFEE156 pKa = 4.14QEE158 pKa = 4.02EE159 pKa = 4.22SDD161 pKa = 3.49EE162 pKa = 4.6YY163 pKa = 11.75YY164 pKa = 11.25DD165 pKa = 4.36FIYY168 pKa = 10.98NFGSSDD174 pKa = 3.8GEE176 pKa = 4.33LVWRR180 pKa = 11.84LNADD184 pKa = 3.39EE185 pKa = 5.01AANEE189 pKa = 4.06FDD191 pKa = 3.99GQLTQEE197 pKa = 5.1DD198 pKa = 4.75IEE200 pKa = 4.42TLKK203 pKa = 10.89GGEE206 pKa = 3.98MSEE209 pKa = 3.98FAQNIEE215 pKa = 3.94VLEE218 pKa = 4.27VEE220 pKa = 4.6KK221 pKa = 10.38VTHH224 pKa = 6.27NGEE227 pKa = 4.24EE228 pKa = 4.23IEE230 pKa = 4.7FPTDD234 pKa = 3.23QEE236 pKa = 4.08IMIEE240 pKa = 4.27SEE242 pKa = 4.19GEE244 pKa = 4.12VYY246 pKa = 10.26IDD248 pKa = 3.41SDD250 pKa = 4.56FITTVADD257 pKa = 3.53YY258 pKa = 10.81EE259 pKa = 4.24LTYY262 pKa = 10.93DD263 pKa = 4.13EE264 pKa = 4.65EE265 pKa = 4.28NQYY268 pKa = 10.89VEE270 pKa = 4.59VIEE273 pKa = 4.84GKK275 pKa = 10.57ADD277 pKa = 3.72FQFEE281 pKa = 4.29PQFDD285 pKa = 4.21EE286 pKa = 4.16NDD288 pKa = 3.47GGIIEE293 pKa = 4.32VNEE296 pKa = 4.5FYY298 pKa = 10.75FDD300 pKa = 3.83EE301 pKa = 4.49NDD303 pKa = 3.43EE304 pKa = 4.45YY305 pKa = 10.86ISRR308 pKa = 11.84SDD310 pKa = 3.57YY311 pKa = 10.6KK312 pKa = 11.22AEE314 pKa = 4.03SQEE317 pKa = 4.04LYY319 pKa = 11.33NLIEE323 pKa = 4.48YY324 pKa = 9.69KK325 pKa = 10.39NHH327 pKa = 6.02VYY329 pKa = 10.67LSYY332 pKa = 11.06AITNDD337 pKa = 3.36FLKK340 pKa = 10.15EE341 pKa = 3.96TVNYY345 pKa = 8.45NQSEE349 pKa = 4.61STVEE353 pKa = 3.31IGEE356 pKa = 4.15NSKK359 pKa = 10.75SKK361 pKa = 10.15QVSEE365 pKa = 4.34ILHH368 pKa = 6.76DD369 pKa = 4.11SGGPKK374 pKa = 10.09AKK376 pKa = 10.49EE377 pKa = 3.6NVTIQGEE384 pKa = 4.52KK385 pKa = 10.03YY386 pKa = 10.51DD387 pKa = 3.91GAIDD391 pKa = 3.93FDD393 pKa = 4.59DD394 pKa = 3.84VGSFTEE400 pKa = 4.3YY401 pKa = 9.58VWLNGEE407 pKa = 4.06HH408 pKa = 7.05KK409 pKa = 10.32YY410 pKa = 10.73SHH412 pKa = 7.04FNGSIYY418 pKa = 10.84YY419 pKa = 8.59KK420 pKa = 9.9TDD422 pKa = 3.1SNLTLTVSFVKK433 pKa = 10.54NDD435 pKa = 3.2NNQTVIKK442 pKa = 9.87EE443 pKa = 4.09FEE445 pKa = 4.3LEE447 pKa = 4.0PGEE450 pKa = 4.27MKK452 pKa = 10.75DD453 pKa = 3.73VDD455 pKa = 3.24IDD457 pKa = 3.64FSGEE461 pKa = 3.43HH462 pKa = 5.48FMGVFSKK469 pKa = 10.66QAIGNNTGGQLLVIGEE485 pKa = 4.34VKK487 pKa = 10.64
MM1 pKa = 7.6KK2 pKa = 10.25KK3 pKa = 9.68ILLIAILFTMSVVIMACSSDD23 pKa = 3.46EE24 pKa = 4.06ADD26 pKa = 3.33SVEE29 pKa = 4.39EE30 pKa = 3.93NVEE33 pKa = 4.05SDD35 pKa = 4.28GEE37 pKa = 4.36TEE39 pKa = 4.04EE40 pKa = 5.55QSDD43 pKa = 3.85GDD45 pKa = 4.13EE46 pKa = 3.92NTLYY50 pKa = 10.93DD51 pKa = 4.09EE52 pKa = 5.32SNLLSLGQSGSIEE65 pKa = 4.23ATNSSYY71 pKa = 11.2EE72 pKa = 3.93VTVEE76 pKa = 3.98SFEE79 pKa = 4.12VLNDD83 pKa = 3.24IVGQQSSEE91 pKa = 3.81EE92 pKa = 4.25VYY94 pKa = 10.7ILLDD98 pKa = 3.61YY99 pKa = 11.06QITNTGEE106 pKa = 3.69EE107 pKa = 4.61AIQGLAVTEE116 pKa = 4.33GNLFNAAGAKK126 pKa = 9.17GLSVTSYY133 pKa = 11.4DD134 pKa = 4.43FISIPDD140 pKa = 3.83TEE142 pKa = 4.39IKK144 pKa = 10.28PGEE147 pKa = 4.15SAAAQILFEE156 pKa = 4.14QEE158 pKa = 4.02EE159 pKa = 4.22SDD161 pKa = 3.49EE162 pKa = 4.6YY163 pKa = 11.75YY164 pKa = 11.25DD165 pKa = 4.36FIYY168 pKa = 10.98NFGSSDD174 pKa = 3.8GEE176 pKa = 4.33LVWRR180 pKa = 11.84LNADD184 pKa = 3.39EE185 pKa = 5.01AANEE189 pKa = 4.06FDD191 pKa = 3.99GQLTQEE197 pKa = 5.1DD198 pKa = 4.75IEE200 pKa = 4.42TLKK203 pKa = 10.89GGEE206 pKa = 3.98MSEE209 pKa = 3.98FAQNIEE215 pKa = 3.94VLEE218 pKa = 4.27VEE220 pKa = 4.6KK221 pKa = 10.38VTHH224 pKa = 6.27NGEE227 pKa = 4.24EE228 pKa = 4.23IEE230 pKa = 4.7FPTDD234 pKa = 3.23QEE236 pKa = 4.08IMIEE240 pKa = 4.27SEE242 pKa = 4.19GEE244 pKa = 4.12VYY246 pKa = 10.26IDD248 pKa = 3.41SDD250 pKa = 4.56FITTVADD257 pKa = 3.53YY258 pKa = 10.81EE259 pKa = 4.24LTYY262 pKa = 10.93DD263 pKa = 4.13EE264 pKa = 4.65EE265 pKa = 4.28NQYY268 pKa = 10.89VEE270 pKa = 4.59VIEE273 pKa = 4.84GKK275 pKa = 10.57ADD277 pKa = 3.72FQFEE281 pKa = 4.29PQFDD285 pKa = 4.21EE286 pKa = 4.16NDD288 pKa = 3.47GGIIEE293 pKa = 4.32VNEE296 pKa = 4.5FYY298 pKa = 10.75FDD300 pKa = 3.83EE301 pKa = 4.49NDD303 pKa = 3.43EE304 pKa = 4.45YY305 pKa = 10.86ISRR308 pKa = 11.84SDD310 pKa = 3.57YY311 pKa = 10.6KK312 pKa = 11.22AEE314 pKa = 4.03SQEE317 pKa = 4.04LYY319 pKa = 11.33NLIEE323 pKa = 4.48YY324 pKa = 9.69KK325 pKa = 10.39NHH327 pKa = 6.02VYY329 pKa = 10.67LSYY332 pKa = 11.06AITNDD337 pKa = 3.36FLKK340 pKa = 10.15EE341 pKa = 3.96TVNYY345 pKa = 8.45NQSEE349 pKa = 4.61STVEE353 pKa = 3.31IGEE356 pKa = 4.15NSKK359 pKa = 10.75SKK361 pKa = 10.15QVSEE365 pKa = 4.34ILHH368 pKa = 6.76DD369 pKa = 4.11SGGPKK374 pKa = 10.09AKK376 pKa = 10.49EE377 pKa = 3.6NVTIQGEE384 pKa = 4.52KK385 pKa = 10.03YY386 pKa = 10.51DD387 pKa = 3.91GAIDD391 pKa = 3.93FDD393 pKa = 4.59DD394 pKa = 3.84VGSFTEE400 pKa = 4.3YY401 pKa = 9.58VWLNGEE407 pKa = 4.06HH408 pKa = 7.05KK409 pKa = 10.32YY410 pKa = 10.73SHH412 pKa = 7.04FNGSIYY418 pKa = 10.84YY419 pKa = 8.59KK420 pKa = 9.9TDD422 pKa = 3.1SNLTLTVSFVKK433 pKa = 10.54NDD435 pKa = 3.2NNQTVIKK442 pKa = 9.87EE443 pKa = 4.09FEE445 pKa = 4.3LEE447 pKa = 4.0PGEE450 pKa = 4.27MKK452 pKa = 10.75DD453 pKa = 3.73VDD455 pKa = 3.24IDD457 pKa = 3.64FSGEE461 pKa = 3.43HH462 pKa = 5.48FMGVFSKK469 pKa = 10.66QAIGNNTGGQLLVIGEE485 pKa = 4.34VKK487 pKa = 10.64
Molecular weight: 54.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4KL54|A0A1I4KL54_9BACI Transcriptional regulator BadM/Rrf2 family OS=Gracilibacillus orientalis OX=334253 GN=SAMN04487943_10414 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSTKK25 pKa = 9.9NGKK28 pKa = 9.88KK29 pKa = 9.59IIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.39KK37 pKa = 10.08GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSTKK25 pKa = 9.9NGKK28 pKa = 9.88KK29 pKa = 9.59IIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.39KK37 pKa = 10.08GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1301525 |
39 |
3190 |
304.2 |
34.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.419 ± 0.037 | 0.633 ± 0.011 |
5.737 ± 0.04 | 7.77 ± 0.059 |
4.596 ± 0.035 | 6.448 ± 0.039 |
2.149 ± 0.02 | 8.131 ± 0.042 |
6.47 ± 0.033 | 9.438 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.726 ± 0.016 | 4.862 ± 0.033 |
3.473 ± 0.022 | 4.161 ± 0.029 |
3.627 ± 0.026 | 6.042 ± 0.032 |
5.584 ± 0.026 | 6.813 ± 0.033 |
1.156 ± 0.016 | 3.764 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
