
Sewage-associated circular DNA virus-35
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
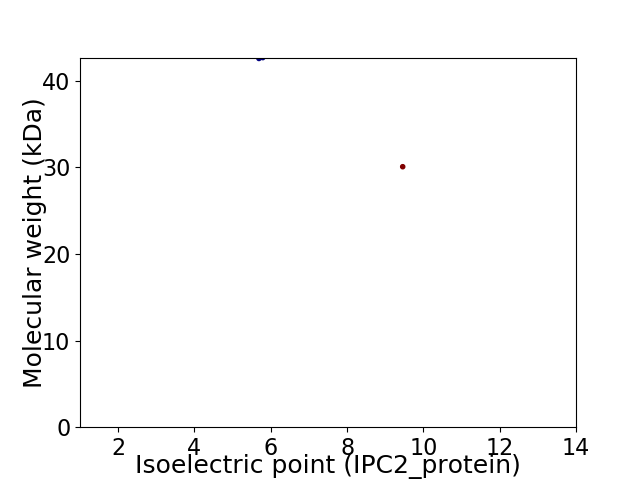
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
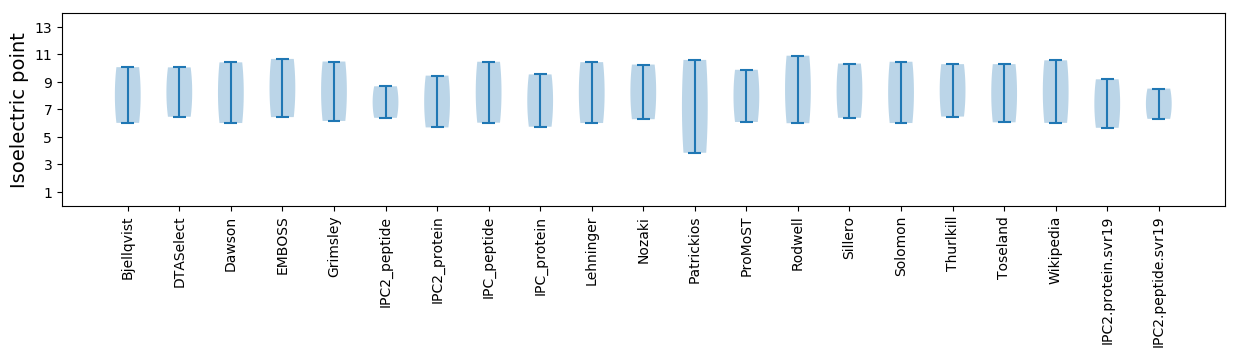
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UI57|A0A0B4UI57_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-35 OX=1592102 PE=4 SV=1
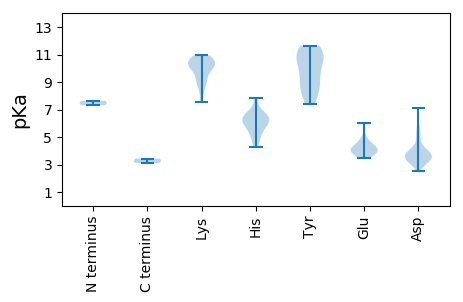
MM1 pKa = 7.63EE2 pKa = 5.59EE3 pKa = 4.04GRR5 pKa = 11.84EE6 pKa = 4.19SITTAKK12 pKa = 10.37HH13 pKa = 5.77FIGTIYY19 pKa = 10.42LPNASPQDD27 pKa = 3.5YY28 pKa = 9.95CAWNGRR34 pKa = 11.84TEE36 pKa = 4.14AAVHH40 pKa = 6.33LFEE43 pKa = 6.02ACVWQHH49 pKa = 6.52EE50 pKa = 4.48ICPDD54 pKa = 3.34TGRR57 pKa = 11.84HH58 pKa = 5.93HH59 pKa = 6.26IQIYY63 pKa = 8.03CHH65 pKa = 6.19ARR67 pKa = 11.84RR68 pKa = 11.84KK69 pKa = 9.51VRR71 pKa = 11.84YY72 pKa = 8.63AQLNDD77 pKa = 3.3ALGVVGSWKK86 pKa = 7.52TCKK89 pKa = 10.57GIAHH93 pKa = 7.83AIACYY98 pKa = 9.88AYY100 pKa = 8.94CTEE103 pKa = 4.35EE104 pKa = 4.61KK105 pKa = 10.44SDD107 pKa = 3.06ASSRR111 pKa = 11.84HH112 pKa = 4.81GCKK115 pKa = 10.08SSGWGDD121 pKa = 3.37PPGRR125 pKa = 11.84GAGGDD130 pKa = 3.47AAISVDD136 pKa = 3.82AAIEE140 pKa = 3.92FAKK143 pKa = 10.37SGKK146 pKa = 8.98RR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84EE150 pKa = 3.54LLMPEE155 pKa = 3.67NAEE158 pKa = 4.19ALRR161 pKa = 11.84GAYY164 pKa = 8.73LHH166 pKa = 6.51PAGFRR171 pKa = 11.84FVSEE175 pKa = 5.0LADD178 pKa = 3.58QPRR181 pKa = 11.84NTPDD185 pKa = 2.56EE186 pKa = 3.87WMQRR190 pKa = 11.84YY191 pKa = 7.75VICFWGGAGVGKK203 pKa = 9.52SRR205 pKa = 11.84RR206 pKa = 11.84VRR208 pKa = 11.84EE209 pKa = 3.78EE210 pKa = 3.49CAAAGLDD217 pKa = 3.68LWVAPASPGGLWFDD231 pKa = 3.89GYY233 pKa = 11.11DD234 pKa = 3.38RR235 pKa = 11.84HH236 pKa = 6.46PCALFDD242 pKa = 5.62DD243 pKa = 5.39FDD245 pKa = 7.09GDD247 pKa = 4.29MPWKK251 pKa = 10.13QLLCILEE258 pKa = 4.18GNIVQVQVKK267 pKa = 8.96GGHH270 pKa = 4.26VAFRR274 pKa = 11.84PRR276 pKa = 11.84VVYY279 pKa = 7.42FTADD283 pKa = 3.81CPPSSWTFSFGKK295 pKa = 10.71GEE297 pKa = 3.89ARR299 pKa = 11.84QGLNQARR306 pKa = 11.84LDD308 pKa = 3.47QLLRR312 pKa = 11.84RR313 pKa = 11.84FCHH316 pKa = 6.73IEE318 pKa = 4.0EE319 pKa = 4.19VSSARR324 pKa = 11.84DD325 pKa = 3.29YY326 pKa = 11.64RR327 pKa = 11.84EE328 pKa = 3.8ALNMNRR334 pKa = 11.84RR335 pKa = 11.84PAGPVPAGGEE345 pKa = 3.84GGANIEE351 pKa = 4.03IAPPLGEE358 pKa = 3.58AWEE361 pKa = 4.33NVHH364 pKa = 6.49VPQLPFDD371 pKa = 3.89VVPDD375 pKa = 3.7SEE377 pKa = 4.83YY378 pKa = 11.57VNEE381 pKa = 4.68LCNQQ385 pKa = 3.44
MM1 pKa = 7.63EE2 pKa = 5.59EE3 pKa = 4.04GRR5 pKa = 11.84EE6 pKa = 4.19SITTAKK12 pKa = 10.37HH13 pKa = 5.77FIGTIYY19 pKa = 10.42LPNASPQDD27 pKa = 3.5YY28 pKa = 9.95CAWNGRR34 pKa = 11.84TEE36 pKa = 4.14AAVHH40 pKa = 6.33LFEE43 pKa = 6.02ACVWQHH49 pKa = 6.52EE50 pKa = 4.48ICPDD54 pKa = 3.34TGRR57 pKa = 11.84HH58 pKa = 5.93HH59 pKa = 6.26IQIYY63 pKa = 8.03CHH65 pKa = 6.19ARR67 pKa = 11.84RR68 pKa = 11.84KK69 pKa = 9.51VRR71 pKa = 11.84YY72 pKa = 8.63AQLNDD77 pKa = 3.3ALGVVGSWKK86 pKa = 7.52TCKK89 pKa = 10.57GIAHH93 pKa = 7.83AIACYY98 pKa = 9.88AYY100 pKa = 8.94CTEE103 pKa = 4.35EE104 pKa = 4.61KK105 pKa = 10.44SDD107 pKa = 3.06ASSRR111 pKa = 11.84HH112 pKa = 4.81GCKK115 pKa = 10.08SSGWGDD121 pKa = 3.37PPGRR125 pKa = 11.84GAGGDD130 pKa = 3.47AAISVDD136 pKa = 3.82AAIEE140 pKa = 3.92FAKK143 pKa = 10.37SGKK146 pKa = 8.98RR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84EE150 pKa = 3.54LLMPEE155 pKa = 3.67NAEE158 pKa = 4.19ALRR161 pKa = 11.84GAYY164 pKa = 8.73LHH166 pKa = 6.51PAGFRR171 pKa = 11.84FVSEE175 pKa = 5.0LADD178 pKa = 3.58QPRR181 pKa = 11.84NTPDD185 pKa = 2.56EE186 pKa = 3.87WMQRR190 pKa = 11.84YY191 pKa = 7.75VICFWGGAGVGKK203 pKa = 9.52SRR205 pKa = 11.84RR206 pKa = 11.84VRR208 pKa = 11.84EE209 pKa = 3.78EE210 pKa = 3.49CAAAGLDD217 pKa = 3.68LWVAPASPGGLWFDD231 pKa = 3.89GYY233 pKa = 11.11DD234 pKa = 3.38RR235 pKa = 11.84HH236 pKa = 6.46PCALFDD242 pKa = 5.62DD243 pKa = 5.39FDD245 pKa = 7.09GDD247 pKa = 4.29MPWKK251 pKa = 10.13QLLCILEE258 pKa = 4.18GNIVQVQVKK267 pKa = 8.96GGHH270 pKa = 4.26VAFRR274 pKa = 11.84PRR276 pKa = 11.84VVYY279 pKa = 7.42FTADD283 pKa = 3.81CPPSSWTFSFGKK295 pKa = 10.71GEE297 pKa = 3.89ARR299 pKa = 11.84QGLNQARR306 pKa = 11.84LDD308 pKa = 3.47QLLRR312 pKa = 11.84RR313 pKa = 11.84FCHH316 pKa = 6.73IEE318 pKa = 4.0EE319 pKa = 4.19VSSARR324 pKa = 11.84DD325 pKa = 3.29YY326 pKa = 11.64RR327 pKa = 11.84EE328 pKa = 3.8ALNMNRR334 pKa = 11.84RR335 pKa = 11.84PAGPVPAGGEE345 pKa = 3.84GGANIEE351 pKa = 4.03IAPPLGEE358 pKa = 3.58AWEE361 pKa = 4.33NVHH364 pKa = 6.49VPQLPFDD371 pKa = 3.89VVPDD375 pKa = 3.7SEE377 pKa = 4.83YY378 pKa = 11.57VNEE381 pKa = 4.68LCNQQ385 pKa = 3.44
Molecular weight: 42.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI57|A0A0B4UI57_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-35 OX=1592102 PE=4 SV=1
MM1 pKa = 7.32SSKK4 pKa = 10.46RR5 pKa = 11.84KK6 pKa = 9.93RR7 pKa = 11.84NGQGEE12 pKa = 4.35GRR14 pKa = 11.84LVFSKK19 pKa = 10.42SRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 10.53GEE25 pKa = 3.63WDD27 pKa = 3.35SYY29 pKa = 10.81VPSSSHH35 pKa = 5.43SAMVAAKK42 pKa = 9.05ARR44 pKa = 11.84QRR46 pKa = 11.84RR47 pKa = 11.84GYY49 pKa = 10.68GSVARR54 pKa = 11.84TRR56 pKa = 11.84GAAVQGEE63 pKa = 4.64TKK65 pKa = 10.92YY66 pKa = 11.01FDD68 pKa = 3.89TEE70 pKa = 4.15LQPTAISAVTTTWVAGTLVDD90 pKa = 3.59PTTTINLGDD99 pKa = 3.84AAIATPGCLFAPKK112 pKa = 10.1VSAALNGRR120 pKa = 11.84IGRR123 pKa = 11.84KK124 pKa = 8.75VLIKK128 pKa = 10.19KK129 pKa = 9.86VKK131 pKa = 7.98LTGVINVPTHH141 pKa = 5.83SAQATGDD148 pKa = 3.58AGTKK152 pKa = 8.92IRR154 pKa = 11.84VMLVLDD160 pKa = 3.62QQTNASAMTSAQLLNDD176 pKa = 4.13AGNALTTISSFQNPNNFGRR195 pKa = 11.84FKK197 pKa = 10.78VLKK200 pKa = 10.31DD201 pKa = 3.03KK202 pKa = 10.93TFFLGDD208 pKa = 3.45LAMTGAATAIEE219 pKa = 4.12QSGTVIPFKK228 pKa = 10.09MSYY231 pKa = 8.87VFKK234 pKa = 10.76RR235 pKa = 11.84GLRR238 pKa = 11.84VDD240 pKa = 5.26FNATNGGTVADD251 pKa = 4.78IIDD254 pKa = 3.82NSLHH258 pKa = 5.22VVCGANNVQLIPSLSYY274 pKa = 9.5YY275 pKa = 10.83SRR277 pKa = 11.84VSYY280 pKa = 11.31VDD282 pKa = 3.12
MM1 pKa = 7.32SSKK4 pKa = 10.46RR5 pKa = 11.84KK6 pKa = 9.93RR7 pKa = 11.84NGQGEE12 pKa = 4.35GRR14 pKa = 11.84LVFSKK19 pKa = 10.42SRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 10.53GEE25 pKa = 3.63WDD27 pKa = 3.35SYY29 pKa = 10.81VPSSSHH35 pKa = 5.43SAMVAAKK42 pKa = 9.05ARR44 pKa = 11.84QRR46 pKa = 11.84RR47 pKa = 11.84GYY49 pKa = 10.68GSVARR54 pKa = 11.84TRR56 pKa = 11.84GAAVQGEE63 pKa = 4.64TKK65 pKa = 10.92YY66 pKa = 11.01FDD68 pKa = 3.89TEE70 pKa = 4.15LQPTAISAVTTTWVAGTLVDD90 pKa = 3.59PTTTINLGDD99 pKa = 3.84AAIATPGCLFAPKK112 pKa = 10.1VSAALNGRR120 pKa = 11.84IGRR123 pKa = 11.84KK124 pKa = 8.75VLIKK128 pKa = 10.19KK129 pKa = 9.86VKK131 pKa = 7.98LTGVINVPTHH141 pKa = 5.83SAQATGDD148 pKa = 3.58AGTKK152 pKa = 8.92IRR154 pKa = 11.84VMLVLDD160 pKa = 3.62QQTNASAMTSAQLLNDD176 pKa = 4.13AGNALTTISSFQNPNNFGRR195 pKa = 11.84FKK197 pKa = 10.78VLKK200 pKa = 10.31DD201 pKa = 3.03KK202 pKa = 10.93TFFLGDD208 pKa = 3.45LAMTGAATAIEE219 pKa = 4.12QSGTVIPFKK228 pKa = 10.09MSYY231 pKa = 8.87VFKK234 pKa = 10.76RR235 pKa = 11.84GLRR238 pKa = 11.84VDD240 pKa = 5.26FNATNGGTVADD251 pKa = 4.78IIDD254 pKa = 3.82NSLHH258 pKa = 5.22VVCGANNVQLIPSLSYY274 pKa = 9.5YY275 pKa = 10.83SRR277 pKa = 11.84VSYY280 pKa = 11.31VDD282 pKa = 3.12
Molecular weight: 30.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
667 |
282 |
385 |
333.5 |
36.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.094 ± 0.067 | 2.549 ± 1.222 |
5.097 ± 0.324 | 4.798 ± 2.009 |
3.898 ± 0.002 | 9.445 ± 0.385 |
2.399 ± 0.887 | 4.348 ± 0.174 |
4.348 ± 1.116 | 6.597 ± 0.329 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.649 ± 0.318 | 4.198 ± 0.745 |
4.948 ± 1.166 | 3.748 ± 0.101 |
6.897 ± 0.577 | 6.447 ± 1.371 |
5.397 ± 2.539 | 7.346 ± 1.009 |
1.949 ± 0.824 | 2.849 ± 0.243 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
