
Wuhan coneheads virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
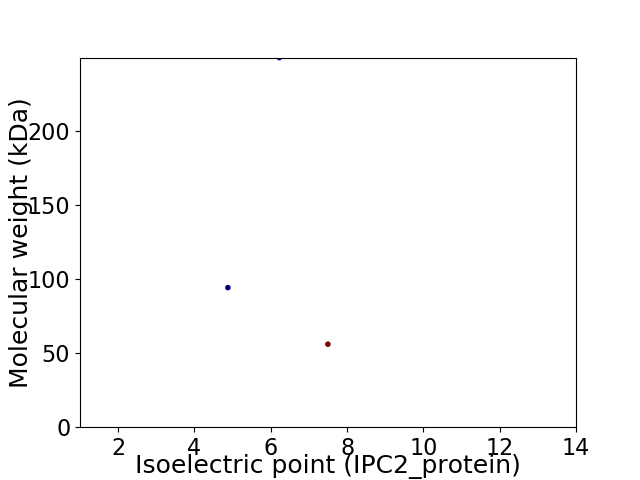
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMM9|A0A1L3KMM9_9VIRU Uncharacterized protein OS=Wuhan coneheads virus 2 OX=1923696 PE=4 SV=1
MM1 pKa = 7.37VSQSASSMPVMAEE14 pKa = 3.81PVLDD18 pKa = 4.02EE19 pKa = 5.23ALTTPTSLGNSTNLATTHH37 pKa = 6.91EE38 pKa = 4.34IANPCAMVVDD48 pKa = 4.28NPMASGRR55 pKa = 11.84PYY57 pKa = 11.28DD58 pKa = 3.52VFTWNRR64 pKa = 11.84KK65 pKa = 8.55HH66 pKa = 5.47YY67 pKa = 10.55QLIKK71 pKa = 10.02SGKK74 pKa = 7.47ITSTDD79 pKa = 2.86TDD81 pKa = 3.84GANLTFVTNNPSEE94 pKa = 4.0FSGLLGAEE102 pKa = 4.2ANLHH106 pKa = 5.61EE107 pKa = 5.11FCTPQIQYY115 pKa = 9.11VVRR118 pKa = 11.84IYY120 pKa = 9.92GTEE123 pKa = 4.22GIIGRR128 pKa = 11.84CLVAAGDD135 pKa = 3.85SSLFISNKK143 pKa = 9.56FEE145 pKa = 4.88AGTGLSDD152 pKa = 4.91ALSAPYY158 pKa = 9.15WRR160 pKa = 11.84YY161 pKa = 9.91ISFNGVSSIEE171 pKa = 3.92FTLNDD176 pKa = 3.07ARR178 pKa = 11.84KK179 pKa = 9.16DD180 pKa = 3.39GFYY183 pKa = 11.0RR184 pKa = 11.84SMTDD188 pKa = 4.94ASSTWQYY195 pKa = 11.62LGIFLVQAPRR205 pKa = 11.84SKK207 pKa = 10.76YY208 pKa = 8.66ATSTATTTDD217 pKa = 3.4VVSLDD222 pKa = 3.19WEE224 pKa = 3.9IFARR228 pKa = 11.84YY229 pKa = 8.73GPRR232 pKa = 11.84HH233 pKa = 5.54MFFGPRR239 pKa = 11.84LVKK242 pKa = 9.32PTTLTLNFTTTLQSFMPAAFDD263 pKa = 3.47FCYY266 pKa = 10.57GSAYY270 pKa = 9.83IPDD273 pKa = 4.57DD274 pKa = 3.71NEE276 pKa = 3.92KK277 pKa = 10.62HH278 pKa = 4.25VTFPTFLQADD288 pKa = 4.4GSLKK292 pKa = 10.38FDD294 pKa = 5.09LSAQSYY300 pKa = 8.52IGSRR304 pKa = 11.84KK305 pKa = 9.48VRR307 pKa = 11.84DD308 pKa = 4.09CYY310 pKa = 10.99TNTIVSVGQVAEE322 pKa = 4.13EE323 pKa = 3.95LDD325 pKa = 4.13IISGASGFNWGVSDD339 pKa = 5.2EE340 pKa = 4.15VMQRR344 pKa = 11.84CMNFGLFQFWGEE356 pKa = 4.44GEE358 pKa = 4.39VSQSLLDD365 pKa = 3.59KK366 pKa = 10.89LDD368 pKa = 4.29NIKK371 pKa = 10.8VLLDD375 pKa = 3.35DD376 pKa = 4.99TFPLMLDD383 pKa = 3.51TPVTFISDD391 pKa = 3.32QEE393 pKa = 3.96YY394 pKa = 10.06DD395 pKa = 3.66YY396 pKa = 11.56SGSEE400 pKa = 3.87LDD402 pKa = 3.68PYY404 pKa = 11.62EE405 pKa = 4.49MIKK408 pKa = 10.64KK409 pKa = 10.0ADD411 pKa = 3.77NKK413 pKa = 10.28QVIQIRR419 pKa = 11.84GCITSGKK426 pKa = 8.17GRR428 pKa = 11.84IWITEE433 pKa = 3.9PSQSHH438 pKa = 5.13VMSSFGDD445 pKa = 3.37VGILYY450 pKa = 9.54FYY452 pKa = 10.96SNKK455 pKa = 10.39NGTKK459 pKa = 9.75RR460 pKa = 11.84PEE462 pKa = 3.7WTSALWGDD470 pKa = 4.12DD471 pKa = 3.23VHH473 pKa = 8.39DD474 pKa = 4.79RR475 pKa = 11.84DD476 pKa = 4.15AGAMAMAYY484 pKa = 10.58YY485 pKa = 10.44EE486 pKa = 4.27DD487 pKa = 4.41AEE489 pKa = 4.69NNVEE493 pKa = 4.07APFIEE498 pKa = 4.64TVSPIISSFKK508 pKa = 10.83LEE510 pKa = 3.97QEE512 pKa = 4.13LDD514 pKa = 3.82DD515 pKa = 5.96KK516 pKa = 11.32VVEE519 pKa = 3.92WTYY522 pKa = 11.39CVNNYY527 pKa = 10.79DD528 pKa = 3.47KK529 pKa = 11.51VKK531 pKa = 10.65FLGPRR536 pKa = 11.84LAEE539 pKa = 4.11LQFKK543 pKa = 10.71RR544 pKa = 11.84PGSSVINKK552 pKa = 8.2SVRR555 pKa = 11.84TTPGFEE561 pKa = 3.98AAGSSDD567 pKa = 3.28FNNYY571 pKa = 9.75LEE573 pKa = 4.81AIRR576 pKa = 11.84RR577 pKa = 11.84QFKK580 pKa = 10.51CDD582 pKa = 4.15LIGYY586 pKa = 6.52TLTINGRR593 pKa = 11.84YY594 pKa = 7.93YY595 pKa = 11.1CDD597 pKa = 2.89VAFYY601 pKa = 10.32KK602 pKa = 10.54GHH604 pKa = 6.36NVVCTDD610 pKa = 3.33TNFQILRR617 pKa = 11.84NMTQEE622 pKa = 3.85IEE624 pKa = 4.3VTNIRR629 pKa = 11.84PIRR632 pKa = 11.84TLGEE636 pKa = 3.83LRR638 pKa = 11.84TASVLPTYY646 pKa = 7.92WTVEE650 pKa = 3.84ATEE653 pKa = 4.47SFFPEE658 pKa = 3.91HH659 pKa = 6.36LKK661 pKa = 10.79EE662 pKa = 4.44FSSQSAGWMIGGGILSGIGSAIGTWQQQQFQEE694 pKa = 4.21ATQKK698 pKa = 10.98RR699 pKa = 11.84NLDD702 pKa = 3.57YY703 pKa = 10.62LTTRR707 pKa = 11.84DD708 pKa = 3.77EE709 pKa = 4.53LNRR712 pKa = 11.84LQQQVLQAQAHH723 pKa = 5.5QYY725 pKa = 7.48QQQGFWQRR733 pKa = 11.84AGVGATAAGGLATNSAEE750 pKa = 4.24TSSTEE755 pKa = 3.81TSKK758 pKa = 11.46DD759 pKa = 3.15STTPNVTPEE768 pKa = 4.2GLSVLQGNQDD778 pKa = 3.35KK779 pKa = 11.38LEE781 pKa = 3.98ASINYY786 pKa = 8.68KK787 pKa = 10.39PLVQVSDD794 pKa = 4.16FNLDD798 pKa = 3.33EE799 pKa = 5.1PYY801 pKa = 9.42VTSSKK806 pKa = 10.41VAQKK810 pKa = 10.29PYY812 pKa = 9.75NGPMHH817 pKa = 7.24TGPPIPPTNVGFKK830 pKa = 10.22LYY832 pKa = 10.7HH833 pKa = 6.76KK834 pKa = 9.98ISNPNTIEE842 pKa = 4.14NEE844 pKa = 3.76ATQAA848 pKa = 3.37
MM1 pKa = 7.37VSQSASSMPVMAEE14 pKa = 3.81PVLDD18 pKa = 4.02EE19 pKa = 5.23ALTTPTSLGNSTNLATTHH37 pKa = 6.91EE38 pKa = 4.34IANPCAMVVDD48 pKa = 4.28NPMASGRR55 pKa = 11.84PYY57 pKa = 11.28DD58 pKa = 3.52VFTWNRR64 pKa = 11.84KK65 pKa = 8.55HH66 pKa = 5.47YY67 pKa = 10.55QLIKK71 pKa = 10.02SGKK74 pKa = 7.47ITSTDD79 pKa = 2.86TDD81 pKa = 3.84GANLTFVTNNPSEE94 pKa = 4.0FSGLLGAEE102 pKa = 4.2ANLHH106 pKa = 5.61EE107 pKa = 5.11FCTPQIQYY115 pKa = 9.11VVRR118 pKa = 11.84IYY120 pKa = 9.92GTEE123 pKa = 4.22GIIGRR128 pKa = 11.84CLVAAGDD135 pKa = 3.85SSLFISNKK143 pKa = 9.56FEE145 pKa = 4.88AGTGLSDD152 pKa = 4.91ALSAPYY158 pKa = 9.15WRR160 pKa = 11.84YY161 pKa = 9.91ISFNGVSSIEE171 pKa = 3.92FTLNDD176 pKa = 3.07ARR178 pKa = 11.84KK179 pKa = 9.16DD180 pKa = 3.39GFYY183 pKa = 11.0RR184 pKa = 11.84SMTDD188 pKa = 4.94ASSTWQYY195 pKa = 11.62LGIFLVQAPRR205 pKa = 11.84SKK207 pKa = 10.76YY208 pKa = 8.66ATSTATTTDD217 pKa = 3.4VVSLDD222 pKa = 3.19WEE224 pKa = 3.9IFARR228 pKa = 11.84YY229 pKa = 8.73GPRR232 pKa = 11.84HH233 pKa = 5.54MFFGPRR239 pKa = 11.84LVKK242 pKa = 9.32PTTLTLNFTTTLQSFMPAAFDD263 pKa = 3.47FCYY266 pKa = 10.57GSAYY270 pKa = 9.83IPDD273 pKa = 4.57DD274 pKa = 3.71NEE276 pKa = 3.92KK277 pKa = 10.62HH278 pKa = 4.25VTFPTFLQADD288 pKa = 4.4GSLKK292 pKa = 10.38FDD294 pKa = 5.09LSAQSYY300 pKa = 8.52IGSRR304 pKa = 11.84KK305 pKa = 9.48VRR307 pKa = 11.84DD308 pKa = 4.09CYY310 pKa = 10.99TNTIVSVGQVAEE322 pKa = 4.13EE323 pKa = 3.95LDD325 pKa = 4.13IISGASGFNWGVSDD339 pKa = 5.2EE340 pKa = 4.15VMQRR344 pKa = 11.84CMNFGLFQFWGEE356 pKa = 4.44GEE358 pKa = 4.39VSQSLLDD365 pKa = 3.59KK366 pKa = 10.89LDD368 pKa = 4.29NIKK371 pKa = 10.8VLLDD375 pKa = 3.35DD376 pKa = 4.99TFPLMLDD383 pKa = 3.51TPVTFISDD391 pKa = 3.32QEE393 pKa = 3.96YY394 pKa = 10.06DD395 pKa = 3.66YY396 pKa = 11.56SGSEE400 pKa = 3.87LDD402 pKa = 3.68PYY404 pKa = 11.62EE405 pKa = 4.49MIKK408 pKa = 10.64KK409 pKa = 10.0ADD411 pKa = 3.77NKK413 pKa = 10.28QVIQIRR419 pKa = 11.84GCITSGKK426 pKa = 8.17GRR428 pKa = 11.84IWITEE433 pKa = 3.9PSQSHH438 pKa = 5.13VMSSFGDD445 pKa = 3.37VGILYY450 pKa = 9.54FYY452 pKa = 10.96SNKK455 pKa = 10.39NGTKK459 pKa = 9.75RR460 pKa = 11.84PEE462 pKa = 3.7WTSALWGDD470 pKa = 4.12DD471 pKa = 3.23VHH473 pKa = 8.39DD474 pKa = 4.79RR475 pKa = 11.84DD476 pKa = 4.15AGAMAMAYY484 pKa = 10.58YY485 pKa = 10.44EE486 pKa = 4.27DD487 pKa = 4.41AEE489 pKa = 4.69NNVEE493 pKa = 4.07APFIEE498 pKa = 4.64TVSPIISSFKK508 pKa = 10.83LEE510 pKa = 3.97QEE512 pKa = 4.13LDD514 pKa = 3.82DD515 pKa = 5.96KK516 pKa = 11.32VVEE519 pKa = 3.92WTYY522 pKa = 11.39CVNNYY527 pKa = 10.79DD528 pKa = 3.47KK529 pKa = 11.51VKK531 pKa = 10.65FLGPRR536 pKa = 11.84LAEE539 pKa = 4.11LQFKK543 pKa = 10.71RR544 pKa = 11.84PGSSVINKK552 pKa = 8.2SVRR555 pKa = 11.84TTPGFEE561 pKa = 3.98AAGSSDD567 pKa = 3.28FNNYY571 pKa = 9.75LEE573 pKa = 4.81AIRR576 pKa = 11.84RR577 pKa = 11.84QFKK580 pKa = 10.51CDD582 pKa = 4.15LIGYY586 pKa = 6.52TLTINGRR593 pKa = 11.84YY594 pKa = 7.93YY595 pKa = 11.1CDD597 pKa = 2.89VAFYY601 pKa = 10.32KK602 pKa = 10.54GHH604 pKa = 6.36NVVCTDD610 pKa = 3.33TNFQILRR617 pKa = 11.84NMTQEE622 pKa = 3.85IEE624 pKa = 4.3VTNIRR629 pKa = 11.84PIRR632 pKa = 11.84TLGEE636 pKa = 3.83LRR638 pKa = 11.84TASVLPTYY646 pKa = 7.92WTVEE650 pKa = 3.84ATEE653 pKa = 4.47SFFPEE658 pKa = 3.91HH659 pKa = 6.36LKK661 pKa = 10.79EE662 pKa = 4.44FSSQSAGWMIGGGILSGIGSAIGTWQQQQFQEE694 pKa = 4.21ATQKK698 pKa = 10.98RR699 pKa = 11.84NLDD702 pKa = 3.57YY703 pKa = 10.62LTTRR707 pKa = 11.84DD708 pKa = 3.77EE709 pKa = 4.53LNRR712 pKa = 11.84LQQQVLQAQAHH723 pKa = 5.5QYY725 pKa = 7.48QQQGFWQRR733 pKa = 11.84AGVGATAAGGLATNSAEE750 pKa = 4.24TSSTEE755 pKa = 3.81TSKK758 pKa = 11.46DD759 pKa = 3.15STTPNVTPEE768 pKa = 4.2GLSVLQGNQDD778 pKa = 3.35KK779 pKa = 11.38LEE781 pKa = 3.98ASINYY786 pKa = 8.68KK787 pKa = 10.39PLVQVSDD794 pKa = 4.16FNLDD798 pKa = 3.33EE799 pKa = 5.1PYY801 pKa = 9.42VTSSKK806 pKa = 10.41VAQKK810 pKa = 10.29PYY812 pKa = 9.75NGPMHH817 pKa = 7.24TGPPIPPTNVGFKK830 pKa = 10.22LYY832 pKa = 10.7HH833 pKa = 6.76KK834 pKa = 9.98ISNPNTIEE842 pKa = 4.14NEE844 pKa = 3.76ATQAA848 pKa = 3.37
Molecular weight: 94.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KM50|A0A1L3KM50_9VIRU Uncharacterized protein OS=Wuhan coneheads virus 2 OX=1923696 PE=4 SV=1
MM1 pKa = 7.61LKK3 pKa = 9.34MEE5 pKa = 4.46TLKK8 pKa = 10.8LSKK11 pKa = 10.69VFTGRR16 pKa = 11.84IFCAEE21 pKa = 4.14DD22 pKa = 3.84LIGVLMEE29 pKa = 4.22RR30 pKa = 11.84KK31 pKa = 9.31FLGEE35 pKa = 3.95LYY37 pKa = 10.59ARR39 pKa = 11.84AMKK42 pKa = 10.1FDD44 pKa = 4.29PYY46 pKa = 11.3VGVGCNPMTGFDD58 pKa = 3.32AYY60 pKa = 10.38FRR62 pKa = 11.84RR63 pKa = 11.84ASPFPHH69 pKa = 6.67RR70 pKa = 11.84FVIDD74 pKa = 3.9FKK76 pKa = 11.24RR77 pKa = 11.84WDD79 pKa = 3.36KK80 pKa = 10.53TLAKK84 pKa = 10.15SLVKK88 pKa = 10.19ILRR91 pKa = 11.84TSLGEE96 pKa = 3.86VNPNLKK102 pKa = 9.89QCFNSVFNSVYY113 pKa = 8.54EE114 pKa = 4.34TYY116 pKa = 10.13HH117 pKa = 6.94ISGTTMYY124 pKa = 10.03YY125 pKa = 10.3RR126 pKa = 11.84SNGQPSGSAGTAVTNSLYY144 pKa = 11.05NLILTYY150 pKa = 10.13TVFSVLYY157 pKa = 10.09KK158 pKa = 10.23EE159 pKa = 5.07QFYY162 pKa = 10.65CYY164 pKa = 7.71PTWEE168 pKa = 4.08EE169 pKa = 3.66FKK171 pKa = 10.91RR172 pKa = 11.84AVQCFFYY179 pKa = 10.99GDD181 pKa = 3.73DD182 pKa = 3.53QFICTVYY189 pKa = 9.65PWFNRR194 pKa = 11.84VTFAAKK200 pKa = 10.11VKK202 pKa = 10.43EE203 pKa = 4.31LFNMNADD210 pKa = 3.84AMDD213 pKa = 3.86KK214 pKa = 10.7AGKK217 pKa = 9.21EE218 pKa = 3.81LSPFDD223 pKa = 3.65EE224 pKa = 4.76WEE226 pKa = 4.09DD227 pKa = 4.09CNWISRR233 pKa = 11.84YY234 pKa = 7.19WRR236 pKa = 11.84KK237 pKa = 9.81LDD239 pKa = 3.51EE240 pKa = 4.29FDD242 pKa = 4.66FRR244 pKa = 11.84VGALKK249 pKa = 10.65KK250 pKa = 10.06MSIGAQFHH258 pKa = 4.94WTSNSEE264 pKa = 3.86ISQIAHH270 pKa = 6.67NIQWALEE277 pKa = 3.95EE278 pKa = 3.87ASIWDD283 pKa = 3.35EE284 pKa = 3.91EE285 pKa = 4.85YY286 pKa = 11.07YY287 pKa = 11.01EE288 pKa = 4.35KK289 pKa = 10.91VKK291 pKa = 10.48QAARR295 pKa = 11.84IILQHH300 pKa = 5.83FPQYY304 pKa = 10.19RR305 pKa = 11.84GEE307 pKa = 4.05FYY309 pKa = 10.49IPSRR313 pKa = 11.84EE314 pKa = 4.0SLQLRR319 pKa = 11.84IYY321 pKa = 11.06NEE323 pKa = 3.63ALTPKK328 pKa = 10.77NLMAQSNEE336 pKa = 3.43AEE338 pKa = 3.85RR339 pKa = 11.84RR340 pKa = 11.84LNSMNIEE347 pKa = 4.09KK348 pKa = 10.73LLVFQPSGKK357 pKa = 8.02TNDD360 pKa = 3.63MASMILNRR368 pKa = 11.84FVQVNDD374 pKa = 3.37VKK376 pKa = 11.29LKK378 pKa = 9.57TDD380 pKa = 3.46YY381 pKa = 10.58TRR383 pKa = 11.84EE384 pKa = 4.45GPQHH388 pKa = 6.94LSMWTCIMTLDD399 pKa = 3.86NYY401 pKa = 10.02RR402 pKa = 11.84VCAEE406 pKa = 3.91AHH408 pKa = 5.57SKK410 pKa = 10.54KK411 pKa = 9.75IARR414 pKa = 11.84EE415 pKa = 3.66MASQKK420 pKa = 9.8MCDD423 pKa = 3.48KK424 pKa = 10.97LGLSLGSKK432 pKa = 10.09FRR434 pKa = 11.84QEE436 pKa = 4.16CDD438 pKa = 3.0AKK440 pKa = 10.72IATLWKK446 pKa = 9.59EE447 pKa = 3.86LARR450 pKa = 11.84LSLVRR455 pKa = 11.84DD456 pKa = 3.4QCSSKK461 pKa = 10.8KK462 pKa = 10.17FGRR465 pKa = 11.84PSYY468 pKa = 10.15EE469 pKa = 4.1DD470 pKa = 3.57SSDD473 pKa = 3.57SSSEE477 pKa = 4.16SEE479 pKa = 4.62DD480 pKa = 3.51FF481 pKa = 4.44
MM1 pKa = 7.61LKK3 pKa = 9.34MEE5 pKa = 4.46TLKK8 pKa = 10.8LSKK11 pKa = 10.69VFTGRR16 pKa = 11.84IFCAEE21 pKa = 4.14DD22 pKa = 3.84LIGVLMEE29 pKa = 4.22RR30 pKa = 11.84KK31 pKa = 9.31FLGEE35 pKa = 3.95LYY37 pKa = 10.59ARR39 pKa = 11.84AMKK42 pKa = 10.1FDD44 pKa = 4.29PYY46 pKa = 11.3VGVGCNPMTGFDD58 pKa = 3.32AYY60 pKa = 10.38FRR62 pKa = 11.84RR63 pKa = 11.84ASPFPHH69 pKa = 6.67RR70 pKa = 11.84FVIDD74 pKa = 3.9FKK76 pKa = 11.24RR77 pKa = 11.84WDD79 pKa = 3.36KK80 pKa = 10.53TLAKK84 pKa = 10.15SLVKK88 pKa = 10.19ILRR91 pKa = 11.84TSLGEE96 pKa = 3.86VNPNLKK102 pKa = 9.89QCFNSVFNSVYY113 pKa = 8.54EE114 pKa = 4.34TYY116 pKa = 10.13HH117 pKa = 6.94ISGTTMYY124 pKa = 10.03YY125 pKa = 10.3RR126 pKa = 11.84SNGQPSGSAGTAVTNSLYY144 pKa = 11.05NLILTYY150 pKa = 10.13TVFSVLYY157 pKa = 10.09KK158 pKa = 10.23EE159 pKa = 5.07QFYY162 pKa = 10.65CYY164 pKa = 7.71PTWEE168 pKa = 4.08EE169 pKa = 3.66FKK171 pKa = 10.91RR172 pKa = 11.84AVQCFFYY179 pKa = 10.99GDD181 pKa = 3.73DD182 pKa = 3.53QFICTVYY189 pKa = 9.65PWFNRR194 pKa = 11.84VTFAAKK200 pKa = 10.11VKK202 pKa = 10.43EE203 pKa = 4.31LFNMNADD210 pKa = 3.84AMDD213 pKa = 3.86KK214 pKa = 10.7AGKK217 pKa = 9.21EE218 pKa = 3.81LSPFDD223 pKa = 3.65EE224 pKa = 4.76WEE226 pKa = 4.09DD227 pKa = 4.09CNWISRR233 pKa = 11.84YY234 pKa = 7.19WRR236 pKa = 11.84KK237 pKa = 9.81LDD239 pKa = 3.51EE240 pKa = 4.29FDD242 pKa = 4.66FRR244 pKa = 11.84VGALKK249 pKa = 10.65KK250 pKa = 10.06MSIGAQFHH258 pKa = 4.94WTSNSEE264 pKa = 3.86ISQIAHH270 pKa = 6.67NIQWALEE277 pKa = 3.95EE278 pKa = 3.87ASIWDD283 pKa = 3.35EE284 pKa = 3.91EE285 pKa = 4.85YY286 pKa = 11.07YY287 pKa = 11.01EE288 pKa = 4.35KK289 pKa = 10.91VKK291 pKa = 10.48QAARR295 pKa = 11.84IILQHH300 pKa = 5.83FPQYY304 pKa = 10.19RR305 pKa = 11.84GEE307 pKa = 4.05FYY309 pKa = 10.49IPSRR313 pKa = 11.84EE314 pKa = 4.0SLQLRR319 pKa = 11.84IYY321 pKa = 11.06NEE323 pKa = 3.63ALTPKK328 pKa = 10.77NLMAQSNEE336 pKa = 3.43AEE338 pKa = 3.85RR339 pKa = 11.84RR340 pKa = 11.84LNSMNIEE347 pKa = 4.09KK348 pKa = 10.73LLVFQPSGKK357 pKa = 8.02TNDD360 pKa = 3.63MASMILNRR368 pKa = 11.84FVQVNDD374 pKa = 3.37VKK376 pKa = 11.29LKK378 pKa = 9.57TDD380 pKa = 3.46YY381 pKa = 10.58TRR383 pKa = 11.84EE384 pKa = 4.45GPQHH388 pKa = 6.94LSMWTCIMTLDD399 pKa = 3.86NYY401 pKa = 10.02RR402 pKa = 11.84VCAEE406 pKa = 3.91AHH408 pKa = 5.57SKK410 pKa = 10.54KK411 pKa = 9.75IARR414 pKa = 11.84EE415 pKa = 3.66MASQKK420 pKa = 9.8MCDD423 pKa = 3.48KK424 pKa = 10.97LGLSLGSKK432 pKa = 10.09FRR434 pKa = 11.84QEE436 pKa = 4.16CDD438 pKa = 3.0AKK440 pKa = 10.72IATLWKK446 pKa = 9.59EE447 pKa = 3.86LARR450 pKa = 11.84LSLVRR455 pKa = 11.84DD456 pKa = 3.4QCSSKK461 pKa = 10.8KK462 pKa = 10.17FGRR465 pKa = 11.84PSYY468 pKa = 10.15EE469 pKa = 4.1DD470 pKa = 3.57SSDD473 pKa = 3.57SSSEE477 pKa = 4.16SEE479 pKa = 4.62DD480 pKa = 3.51FF481 pKa = 4.44
Molecular weight: 56.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3522 |
481 |
2193 |
1174.0 |
133.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.735 ± 0.576 | 1.76 ± 0.196 |
4.656 ± 0.422 | 6.871 ± 0.464 |
5.139 ± 0.379 | 5.764 ± 0.485 |
1.988 ± 0.311 | 6.133 ± 0.644 |
6.218 ± 0.679 | 7.865 ± 0.182 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.47 ± 0.266 | 5.196 ± 0.104 |
4.969 ± 0.574 | 4.514 ± 0.222 |
4.373 ± 0.362 | 7.325 ± 0.653 |
6.502 ± 0.658 | 6.871 ± 0.704 |
1.533 ± 0.237 | 4.117 ± 0.19 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
