
Lysobacter enzymogenes
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Lysobacter
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
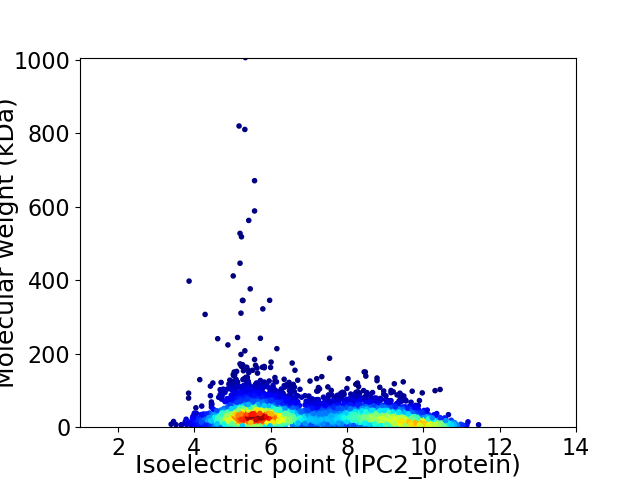
Virtual 2D-PAGE plot for 5514 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2DG49|A0A0S2DG49_LYSEN Uncharacterized protein OS=Lysobacter enzymogenes OX=69 GN=GLE_2131 PE=4 SV=1
MM1 pKa = 7.37GAATDD6 pKa = 3.66IEE8 pKa = 4.58QINLSNPTATGDD20 pKa = 3.93DD21 pKa = 3.79ANGTDD26 pKa = 5.24DD27 pKa = 4.2EE28 pKa = 5.17DD29 pKa = 4.67AFASIPAIPLVAGGTYY45 pKa = 10.41NLSVPCAGNGAAVSGFIDD63 pKa = 4.15FNRR66 pKa = 11.84DD67 pKa = 2.15GDD69 pKa = 4.06FADD72 pKa = 4.12SGEE75 pKa = 4.13KK76 pKa = 10.07SAAATCNGSTAALTWTLPGAAGLNAGASFVRR107 pKa = 11.84LRR109 pKa = 11.84IGTQAAQVNVPTGLASDD126 pKa = 4.86GEE128 pKa = 4.5VEE130 pKa = 4.81DD131 pKa = 4.23YY132 pKa = 11.32AVTLTSPTLTLRR144 pKa = 11.84KK145 pKa = 8.75QWNGATTGDD154 pKa = 4.09DD155 pKa = 3.57AAVTASRR162 pKa = 11.84GGTVVGTLNSDD173 pKa = 3.18AGSANEE179 pKa = 5.0LDD181 pKa = 3.37TATAINVFPGEE192 pKa = 4.27TLTLAEE198 pKa = 4.22TLAANGGRR206 pKa = 11.84TYY208 pKa = 9.84TQSLACTGTSDD219 pKa = 5.1ANPNDD224 pKa = 3.52GLTIGAADD232 pKa = 3.86ANIVCTFTNAQAARR246 pKa = 11.84LTVIKK251 pKa = 10.75NVINDD256 pKa = 3.63NGGVATVADD265 pKa = 4.54FGIQVDD271 pKa = 4.65GVARR275 pKa = 11.84SFGANTGTAGNAVYY289 pKa = 9.7TSAAVAVNAGTRR301 pKa = 11.84ALTEE305 pKa = 4.41LNVAGYY311 pKa = 8.28TEE313 pKa = 4.57GTWACTGTGVTMGNAAFDD331 pKa = 3.99SGSVTLAAGADD342 pKa = 3.74ATCTITNDD350 pKa = 3.49DD351 pKa = 4.07TPASLTLRR359 pKa = 11.84KK360 pKa = 9.72AVVNDD365 pKa = 3.55NGGTAAQTAWTLNAAGPTPISGTHH389 pKa = 5.7GAAAVTNAAVGAGTYY404 pKa = 8.02TLSEE408 pKa = 4.23SGGPAGYY415 pKa = 7.42TASAYY420 pKa = 10.34SCSVDD425 pKa = 2.77GGAAVSGNSLTLANGQNAVCTITNDD450 pKa = 3.55DD451 pKa = 4.37APAQLTLQKK460 pKa = 10.89VVVNDD465 pKa = 3.39NGGAAVATAWTLNANGPTPISGTHH489 pKa = 5.92AAAAVTNATVGAGTYY504 pKa = 7.95TLSEE508 pKa = 4.23SGGPAGYY515 pKa = 7.42TASAYY520 pKa = 10.47SCSIDD525 pKa = 2.97GGAAVSGNSLTLANGQNAVCTITNDD550 pKa = 3.58DD551 pKa = 3.81APANLTLRR559 pKa = 11.84KK560 pKa = 7.83TVVNNNGGTAAATAWTLNAAGPTPISGTHH589 pKa = 6.13GSAAVTNAAVGAGTYY604 pKa = 7.9TLSEE608 pKa = 4.25TGPAGYY614 pKa = 9.41NASAWVCTNGVTVTGGNQITLANGATTDD642 pKa = 3.74CTITNDD648 pKa = 3.19DD649 pKa = 3.73RR650 pKa = 11.84QAVLSLTKK658 pKa = 10.3RR659 pKa = 11.84VINDD663 pKa = 3.22NGGTRR668 pKa = 11.84LASAWTLRR676 pKa = 11.84ATGPTAVSGTPPVLPRR692 pKa = 11.84PVNAGTYY699 pKa = 7.55TLSEE703 pKa = 4.29TGPGGYY709 pKa = 9.53AASAWTCTNGVTVTGGNQITLPPGGGTSCQITNDD743 pKa = 3.95DD744 pKa = 3.98RR745 pKa = 11.84PATLTLQKK753 pKa = 10.36IVVNDD758 pKa = 3.6NGGTAVATAWTLNAAGPTPISGAHH782 pKa = 5.88GAAAVTGAAVNAGTYY797 pKa = 10.42ALSEE801 pKa = 4.34SGGPADD807 pKa = 4.12YY808 pKa = 8.54TASAYY813 pKa = 10.55SCSIDD818 pKa = 2.97GGAAVSGDD826 pKa = 3.94SLTLANGQSAVCTVTNDD843 pKa = 4.29DD844 pKa = 4.39SNQADD849 pKa = 3.82LSITKK854 pKa = 10.49SNTYY858 pKa = 9.41TPADD862 pKa = 3.91PSDD865 pKa = 3.58QVGDD869 pKa = 3.82TVVAGTPTTYY879 pKa = 10.79TLVVTNNGPATAVGAVVRR897 pKa = 11.84DD898 pKa = 3.95VPQAGLDD905 pKa = 3.88CPASNPVACSGAACPSAAITIGDD928 pKa = 4.22LGSGITLGSLATGATATLSFTCAVQQQ954 pKa = 3.9
MM1 pKa = 7.37GAATDD6 pKa = 3.66IEE8 pKa = 4.58QINLSNPTATGDD20 pKa = 3.93DD21 pKa = 3.79ANGTDD26 pKa = 5.24DD27 pKa = 4.2EE28 pKa = 5.17DD29 pKa = 4.67AFASIPAIPLVAGGTYY45 pKa = 10.41NLSVPCAGNGAAVSGFIDD63 pKa = 4.15FNRR66 pKa = 11.84DD67 pKa = 2.15GDD69 pKa = 4.06FADD72 pKa = 4.12SGEE75 pKa = 4.13KK76 pKa = 10.07SAAATCNGSTAALTWTLPGAAGLNAGASFVRR107 pKa = 11.84LRR109 pKa = 11.84IGTQAAQVNVPTGLASDD126 pKa = 4.86GEE128 pKa = 4.5VEE130 pKa = 4.81DD131 pKa = 4.23YY132 pKa = 11.32AVTLTSPTLTLRR144 pKa = 11.84KK145 pKa = 8.75QWNGATTGDD154 pKa = 4.09DD155 pKa = 3.57AAVTASRR162 pKa = 11.84GGTVVGTLNSDD173 pKa = 3.18AGSANEE179 pKa = 5.0LDD181 pKa = 3.37TATAINVFPGEE192 pKa = 4.27TLTLAEE198 pKa = 4.22TLAANGGRR206 pKa = 11.84TYY208 pKa = 9.84TQSLACTGTSDD219 pKa = 5.1ANPNDD224 pKa = 3.52GLTIGAADD232 pKa = 3.86ANIVCTFTNAQAARR246 pKa = 11.84LTVIKK251 pKa = 10.75NVINDD256 pKa = 3.63NGGVATVADD265 pKa = 4.54FGIQVDD271 pKa = 4.65GVARR275 pKa = 11.84SFGANTGTAGNAVYY289 pKa = 9.7TSAAVAVNAGTRR301 pKa = 11.84ALTEE305 pKa = 4.41LNVAGYY311 pKa = 8.28TEE313 pKa = 4.57GTWACTGTGVTMGNAAFDD331 pKa = 3.99SGSVTLAAGADD342 pKa = 3.74ATCTITNDD350 pKa = 3.49DD351 pKa = 4.07TPASLTLRR359 pKa = 11.84KK360 pKa = 9.72AVVNDD365 pKa = 3.55NGGTAAQTAWTLNAAGPTPISGTHH389 pKa = 5.7GAAAVTNAAVGAGTYY404 pKa = 8.02TLSEE408 pKa = 4.23SGGPAGYY415 pKa = 7.42TASAYY420 pKa = 10.34SCSVDD425 pKa = 2.77GGAAVSGNSLTLANGQNAVCTITNDD450 pKa = 3.55DD451 pKa = 4.37APAQLTLQKK460 pKa = 10.89VVVNDD465 pKa = 3.39NGGAAVATAWTLNANGPTPISGTHH489 pKa = 5.92AAAAVTNATVGAGTYY504 pKa = 7.95TLSEE508 pKa = 4.23SGGPAGYY515 pKa = 7.42TASAYY520 pKa = 10.47SCSIDD525 pKa = 2.97GGAAVSGNSLTLANGQNAVCTITNDD550 pKa = 3.58DD551 pKa = 3.81APANLTLRR559 pKa = 11.84KK560 pKa = 7.83TVVNNNGGTAAATAWTLNAAGPTPISGTHH589 pKa = 6.13GSAAVTNAAVGAGTYY604 pKa = 7.9TLSEE608 pKa = 4.25TGPAGYY614 pKa = 9.41NASAWVCTNGVTVTGGNQITLANGATTDD642 pKa = 3.74CTITNDD648 pKa = 3.19DD649 pKa = 3.73RR650 pKa = 11.84QAVLSLTKK658 pKa = 10.3RR659 pKa = 11.84VINDD663 pKa = 3.22NGGTRR668 pKa = 11.84LASAWTLRR676 pKa = 11.84ATGPTAVSGTPPVLPRR692 pKa = 11.84PVNAGTYY699 pKa = 7.55TLSEE703 pKa = 4.29TGPGGYY709 pKa = 9.53AASAWTCTNGVTVTGGNQITLPPGGGTSCQITNDD743 pKa = 3.95DD744 pKa = 3.98RR745 pKa = 11.84PATLTLQKK753 pKa = 10.36IVVNDD758 pKa = 3.6NGGTAVATAWTLNAAGPTPISGAHH782 pKa = 5.88GAAAVTGAAVNAGTYY797 pKa = 10.42ALSEE801 pKa = 4.34SGGPADD807 pKa = 4.12YY808 pKa = 8.54TASAYY813 pKa = 10.55SCSIDD818 pKa = 2.97GGAAVSGDD826 pKa = 3.94SLTLANGQSAVCTVTNDD843 pKa = 4.29DD844 pKa = 4.39SNQADD849 pKa = 3.82LSITKK854 pKa = 10.49SNTYY858 pKa = 9.41TPADD862 pKa = 3.91PSDD865 pKa = 3.58QVGDD869 pKa = 3.82TVVAGTPTTYY879 pKa = 10.79TLVVTNNGPATAVGAVVRR897 pKa = 11.84DD898 pKa = 3.95VPQAGLDD905 pKa = 3.88CPASNPVACSGAACPSAAITIGDD928 pKa = 4.22LGSGITLGSLATGATATLSFTCAVQQQ954 pKa = 3.9
Molecular weight: 92.72 kDa
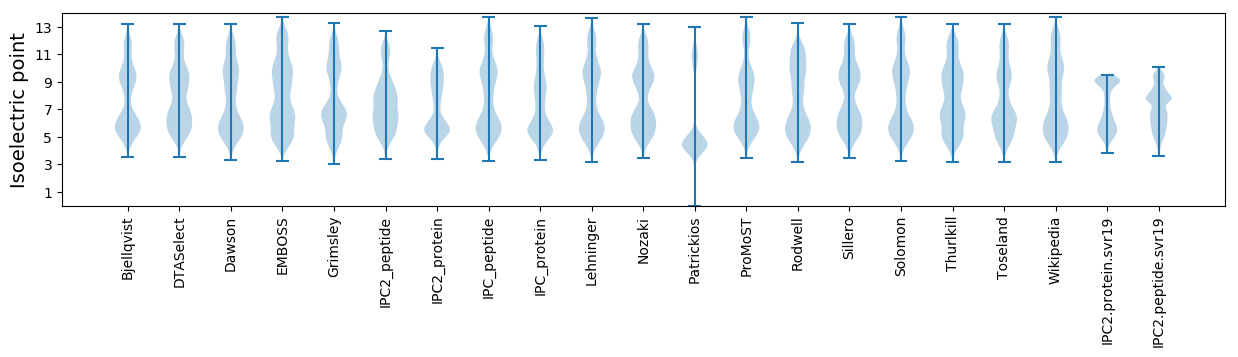
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2DMV5|A0A0S2DMV5_LYSEN Aminopeptidase N OS=Lysobacter enzymogenes OX=69 GN=lta4h PE=3 SV=1
MM1 pKa = 7.13QRR3 pKa = 11.84RR4 pKa = 11.84SPHH7 pKa = 5.79GPAGTGGRR15 pKa = 11.84ARR17 pKa = 11.84PARR20 pKa = 11.84GAPRR24 pKa = 11.84MPRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84GFRR33 pKa = 11.84ARR35 pKa = 11.84AAPARR40 pKa = 11.84AFARR44 pKa = 11.84FGTVFRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84GAAPPPSS60 pKa = 3.51
MM1 pKa = 7.13QRR3 pKa = 11.84RR4 pKa = 11.84SPHH7 pKa = 5.79GPAGTGGRR15 pKa = 11.84ARR17 pKa = 11.84PARR20 pKa = 11.84GAPRR24 pKa = 11.84MPRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84GFRR33 pKa = 11.84ARR35 pKa = 11.84AAPARR40 pKa = 11.84AFARR44 pKa = 11.84FGTVFRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84GAAPPPSS60 pKa = 3.51
Molecular weight: 6.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1754100 |
37 |
9289 |
318.1 |
34.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.471 ± 0.062 | 0.893 ± 0.012 |
5.916 ± 0.032 | 5.13 ± 0.036 |
3.276 ± 0.023 | 8.791 ± 0.05 |
2.065 ± 0.019 | 3.684 ± 0.026 |
2.592 ± 0.037 | 10.5 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.843 ± 0.018 | 2.46 ± 0.029 |
5.626 ± 0.032 | 3.764 ± 0.026 |
8.361 ± 0.048 | 5.222 ± 0.027 |
4.488 ± 0.056 | 6.805 ± 0.028 |
1.556 ± 0.016 | 2.452 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
