
Faeces associated gemycircularvirus 7
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus malas1; Mallard associated gemycircularvirus 1
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
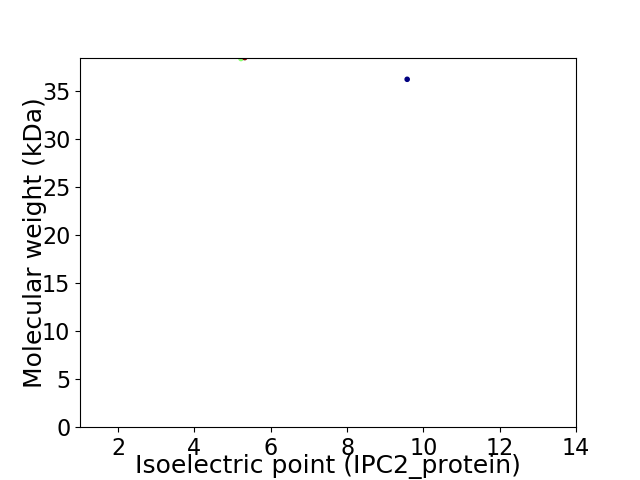
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T1YRX6|T1YRX6_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 7 OX=1391036 PE=3 SV=1
MM1 pKa = 7.64SRR3 pKa = 11.84QFCLKK8 pKa = 10.02DD9 pKa = 3.36AKK11 pKa = 10.79YY12 pKa = 10.93CLLTYY17 pKa = 9.15PQIPEE22 pKa = 3.8TEE24 pKa = 4.1AYY26 pKa = 7.96EE27 pKa = 4.39FPEE30 pKa = 4.29LASEE34 pKa = 4.49LFSSEE39 pKa = 4.45RR40 pKa = 11.84VDD42 pKa = 3.25ANYY45 pKa = 10.53VIGRR49 pKa = 11.84EE50 pKa = 3.9LHH52 pKa = 6.73ADD54 pKa = 3.36GGYY57 pKa = 9.17TSHH60 pKa = 7.21CFLDD64 pKa = 4.41FGRR67 pKa = 11.84KK68 pKa = 8.49FSSRR72 pKa = 11.84DD73 pKa = 3.11TRR75 pKa = 11.84IFDD78 pKa = 3.7IQGHH82 pKa = 5.87HH83 pKa = 6.82PNIEE87 pKa = 4.16RR88 pKa = 11.84VGRR91 pKa = 11.84TPRR94 pKa = 11.84TAYY97 pKa = 10.8NYY99 pKa = 7.81TIKK102 pKa = 11.17DD103 pKa = 3.28NDD105 pKa = 3.32VVARR109 pKa = 11.84SGEE112 pKa = 4.08YY113 pKa = 10.09SGPPEE118 pKa = 4.15NNNSRR123 pKa = 11.84EE124 pKa = 4.05RR125 pKa = 11.84QSGNDD130 pKa = 3.61TSSDD134 pKa = 2.97WATILCAEE142 pKa = 4.39SRR144 pKa = 11.84DD145 pKa = 4.06EE146 pKa = 6.23FFDD149 pKa = 3.72LCKK152 pKa = 10.47SLQPRR157 pKa = 11.84SLACSFLSLTRR168 pKa = 11.84YY169 pKa = 10.08ADD171 pKa = 2.87WRR173 pKa = 11.84YY174 pKa = 10.14RR175 pKa = 11.84PVPTPYY181 pKa = 10.07QHH183 pKa = 7.49PDD185 pKa = 2.69DD186 pKa = 3.67WSFNLEE192 pKa = 4.11SHH194 pKa = 6.62SVLLDD199 pKa = 3.31WVDD202 pKa = 3.17EE203 pKa = 4.28SLRR206 pKa = 11.84GGQDD210 pKa = 2.55RR211 pKa = 11.84SLVMYY216 pKa = 10.89GEE218 pKa = 4.24TRR220 pKa = 11.84LGKK223 pKa = 7.62TVWARR228 pKa = 11.84SLGPHH233 pKa = 7.21LYY235 pKa = 10.24FCGLYY240 pKa = 9.92SYY242 pKa = 11.19KK243 pKa = 10.15EE244 pKa = 3.72ASRR247 pKa = 11.84AHH249 pKa = 5.4EE250 pKa = 4.08AEE252 pKa = 4.0YY253 pKa = 10.54AIFDD257 pKa = 4.38DD258 pKa = 4.32LQGGIKK264 pKa = 10.08FFHH267 pKa = 6.68GFKK270 pKa = 10.25NWLGAQQEE278 pKa = 4.28FQIKK282 pKa = 9.86GLYY285 pKa = 9.46RR286 pKa = 11.84DD287 pKa = 4.08PEE289 pKa = 4.16LLKK292 pKa = 10.23WGKK295 pKa = 9.65PSIWCSNTDD304 pKa = 3.12PRR306 pKa = 11.84QDD308 pKa = 3.41LDD310 pKa = 3.7YY311 pKa = 11.5SDD313 pKa = 5.27RR314 pKa = 11.84CWLEE318 pKa = 3.95GNCVFVEE325 pKa = 4.48VTTSLIGG332 pKa = 3.4
MM1 pKa = 7.64SRR3 pKa = 11.84QFCLKK8 pKa = 10.02DD9 pKa = 3.36AKK11 pKa = 10.79YY12 pKa = 10.93CLLTYY17 pKa = 9.15PQIPEE22 pKa = 3.8TEE24 pKa = 4.1AYY26 pKa = 7.96EE27 pKa = 4.39FPEE30 pKa = 4.29LASEE34 pKa = 4.49LFSSEE39 pKa = 4.45RR40 pKa = 11.84VDD42 pKa = 3.25ANYY45 pKa = 10.53VIGRR49 pKa = 11.84EE50 pKa = 3.9LHH52 pKa = 6.73ADD54 pKa = 3.36GGYY57 pKa = 9.17TSHH60 pKa = 7.21CFLDD64 pKa = 4.41FGRR67 pKa = 11.84KK68 pKa = 8.49FSSRR72 pKa = 11.84DD73 pKa = 3.11TRR75 pKa = 11.84IFDD78 pKa = 3.7IQGHH82 pKa = 5.87HH83 pKa = 6.82PNIEE87 pKa = 4.16RR88 pKa = 11.84VGRR91 pKa = 11.84TPRR94 pKa = 11.84TAYY97 pKa = 10.8NYY99 pKa = 7.81TIKK102 pKa = 11.17DD103 pKa = 3.28NDD105 pKa = 3.32VVARR109 pKa = 11.84SGEE112 pKa = 4.08YY113 pKa = 10.09SGPPEE118 pKa = 4.15NNNSRR123 pKa = 11.84EE124 pKa = 4.05RR125 pKa = 11.84QSGNDD130 pKa = 3.61TSSDD134 pKa = 2.97WATILCAEE142 pKa = 4.39SRR144 pKa = 11.84DD145 pKa = 4.06EE146 pKa = 6.23FFDD149 pKa = 3.72LCKK152 pKa = 10.47SLQPRR157 pKa = 11.84SLACSFLSLTRR168 pKa = 11.84YY169 pKa = 10.08ADD171 pKa = 2.87WRR173 pKa = 11.84YY174 pKa = 10.14RR175 pKa = 11.84PVPTPYY181 pKa = 10.07QHH183 pKa = 7.49PDD185 pKa = 2.69DD186 pKa = 3.67WSFNLEE192 pKa = 4.11SHH194 pKa = 6.62SVLLDD199 pKa = 3.31WVDD202 pKa = 3.17EE203 pKa = 4.28SLRR206 pKa = 11.84GGQDD210 pKa = 2.55RR211 pKa = 11.84SLVMYY216 pKa = 10.89GEE218 pKa = 4.24TRR220 pKa = 11.84LGKK223 pKa = 7.62TVWARR228 pKa = 11.84SLGPHH233 pKa = 7.21LYY235 pKa = 10.24FCGLYY240 pKa = 9.92SYY242 pKa = 11.19KK243 pKa = 10.15EE244 pKa = 3.72ASRR247 pKa = 11.84AHH249 pKa = 5.4EE250 pKa = 4.08AEE252 pKa = 4.0YY253 pKa = 10.54AIFDD257 pKa = 4.38DD258 pKa = 4.32LQGGIKK264 pKa = 10.08FFHH267 pKa = 6.68GFKK270 pKa = 10.25NWLGAQQEE278 pKa = 4.28FQIKK282 pKa = 9.86GLYY285 pKa = 9.46RR286 pKa = 11.84DD287 pKa = 4.08PEE289 pKa = 4.16LLKK292 pKa = 10.23WGKK295 pKa = 9.65PSIWCSNTDD304 pKa = 3.12PRR306 pKa = 11.84QDD308 pKa = 3.41LDD310 pKa = 3.7YY311 pKa = 11.5SDD313 pKa = 5.27RR314 pKa = 11.84CWLEE318 pKa = 3.95GNCVFVEE325 pKa = 4.48VTTSLIGG332 pKa = 3.4
Molecular weight: 38.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1YRX6|T1YRX6_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 7 OX=1391036 PE=3 SV=1
MM1 pKa = 7.78AYY3 pKa = 8.88TRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 6.31GTRR11 pKa = 11.84YY12 pKa = 8.91ARR14 pKa = 11.84RR15 pKa = 11.84GVRR18 pKa = 11.84RR19 pKa = 11.84GSRR22 pKa = 11.84RR23 pKa = 11.84TSTRR27 pKa = 11.84GYY29 pKa = 8.82VRR31 pKa = 11.84KK32 pKa = 9.58RR33 pKa = 11.84ANKK36 pKa = 7.65YY37 pKa = 8.0RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84TGVSRR47 pKa = 11.84KK48 pKa = 9.58RR49 pKa = 11.84ILNITSTKK57 pKa = 10.27KK58 pKa = 10.08ADD60 pKa = 3.47HH61 pKa = 6.8MINANKK67 pKa = 9.67DD68 pKa = 3.65DD69 pKa = 4.41PVTPVNPLSGLGPTIVNPANGVTQCYY95 pKa = 9.78AWIATARR102 pKa = 11.84DD103 pKa = 3.61NTNTPGVPAGGVDD116 pKa = 3.64DD117 pKa = 4.38SSRR120 pKa = 11.84TADD123 pKa = 3.17VCYY126 pKa = 10.14MRR128 pKa = 11.84GLKK131 pKa = 9.77EE132 pKa = 4.13RR133 pKa = 11.84IGLEE137 pKa = 3.99SSSNLPFQWRR147 pKa = 11.84RR148 pKa = 11.84ICFTFKK154 pKa = 10.46GQEE157 pKa = 3.73ILRR160 pKa = 11.84DD161 pKa = 3.65RR162 pKa = 11.84TSTAVGSLYY171 pKa = 10.62QEE173 pKa = 4.44AAPPGWMRR181 pKa = 11.84TTTHH185 pKa = 5.51IQGLNVTSNFVYY197 pKa = 10.81DD198 pKa = 3.83NLEE201 pKa = 4.27TILFRR206 pKa = 11.84GVKK209 pKa = 9.78NLDD212 pKa = 3.18WFDD215 pKa = 3.64EE216 pKa = 4.24MNAQVDD222 pKa = 4.49RR223 pKa = 11.84YY224 pKa = 10.13RR225 pKa = 11.84VDD227 pKa = 3.44LKK229 pKa = 10.8YY230 pKa = 11.17DD231 pKa = 4.45RR232 pKa = 11.84IRR234 pKa = 11.84SFQSGNDD241 pKa = 3.12VGLLRR246 pKa = 11.84KK247 pKa = 9.74INMWHH252 pKa = 6.83PMNKK256 pKa = 9.21NLEE259 pKa = 4.31YY260 pKa = 11.07NSDD263 pKa = 3.34EE264 pKa = 4.41SAGGRR269 pKa = 11.84LEE271 pKa = 4.74SVLSSNTKK279 pKa = 10.52GSMGDD284 pKa = 3.62YY285 pKa = 10.66YY286 pKa = 11.41VIDD289 pKa = 3.82YY290 pKa = 10.52FRR292 pKa = 11.84FSAFATINDD301 pKa = 5.0AIAFTPEE308 pKa = 3.24ACLYY312 pKa = 7.41WHH314 pKa = 6.72EE315 pKa = 4.24RR316 pKa = 3.51
MM1 pKa = 7.78AYY3 pKa = 8.88TRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 6.31GTRR11 pKa = 11.84YY12 pKa = 8.91ARR14 pKa = 11.84RR15 pKa = 11.84GVRR18 pKa = 11.84RR19 pKa = 11.84GSRR22 pKa = 11.84RR23 pKa = 11.84TSTRR27 pKa = 11.84GYY29 pKa = 8.82VRR31 pKa = 11.84KK32 pKa = 9.58RR33 pKa = 11.84ANKK36 pKa = 7.65YY37 pKa = 8.0RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84TGVSRR47 pKa = 11.84KK48 pKa = 9.58RR49 pKa = 11.84ILNITSTKK57 pKa = 10.27KK58 pKa = 10.08ADD60 pKa = 3.47HH61 pKa = 6.8MINANKK67 pKa = 9.67DD68 pKa = 3.65DD69 pKa = 4.41PVTPVNPLSGLGPTIVNPANGVTQCYY95 pKa = 9.78AWIATARR102 pKa = 11.84DD103 pKa = 3.61NTNTPGVPAGGVDD116 pKa = 3.64DD117 pKa = 4.38SSRR120 pKa = 11.84TADD123 pKa = 3.17VCYY126 pKa = 10.14MRR128 pKa = 11.84GLKK131 pKa = 9.77EE132 pKa = 4.13RR133 pKa = 11.84IGLEE137 pKa = 3.99SSSNLPFQWRR147 pKa = 11.84RR148 pKa = 11.84ICFTFKK154 pKa = 10.46GQEE157 pKa = 3.73ILRR160 pKa = 11.84DD161 pKa = 3.65RR162 pKa = 11.84TSTAVGSLYY171 pKa = 10.62QEE173 pKa = 4.44AAPPGWMRR181 pKa = 11.84TTTHH185 pKa = 5.51IQGLNVTSNFVYY197 pKa = 10.81DD198 pKa = 3.83NLEE201 pKa = 4.27TILFRR206 pKa = 11.84GVKK209 pKa = 9.78NLDD212 pKa = 3.18WFDD215 pKa = 3.64EE216 pKa = 4.24MNAQVDD222 pKa = 4.49RR223 pKa = 11.84YY224 pKa = 10.13RR225 pKa = 11.84VDD227 pKa = 3.44LKK229 pKa = 10.8YY230 pKa = 11.17DD231 pKa = 4.45RR232 pKa = 11.84IRR234 pKa = 11.84SFQSGNDD241 pKa = 3.12VGLLRR246 pKa = 11.84KK247 pKa = 9.74INMWHH252 pKa = 6.83PMNKK256 pKa = 9.21NLEE259 pKa = 4.31YY260 pKa = 11.07NSDD263 pKa = 3.34EE264 pKa = 4.41SAGGRR269 pKa = 11.84LEE271 pKa = 4.74SVLSSNTKK279 pKa = 10.52GSMGDD284 pKa = 3.62YY285 pKa = 10.66YY286 pKa = 11.41VIDD289 pKa = 3.82YY290 pKa = 10.52FRR292 pKa = 11.84FSAFATINDD301 pKa = 5.0AIAFTPEE308 pKa = 3.24ACLYY312 pKa = 7.41WHH314 pKa = 6.72EE315 pKa = 4.24RR316 pKa = 3.51
Molecular weight: 36.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
648 |
316 |
332 |
324.0 |
37.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.864 ± 0.548 | 2.16 ± 0.628 |
6.79 ± 0.546 | 5.247 ± 1.24 |
4.475 ± 0.698 | 7.253 ± 0.24 |
2.006 ± 0.52 | 4.167 ± 0.407 |
3.858 ± 0.18 | 7.562 ± 1.087 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.543 ± 0.694 | 5.247 ± 1.204 |
4.321 ± 0.367 | 2.932 ± 0.503 |
9.722 ± 1.617 | 8.025 ± 0.746 |
6.327 ± 1.112 | 4.938 ± 0.754 |
2.315 ± 0.292 | 5.247 ± 0.129 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
