
Sauropus yellowing virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 8.32
Get precalculated fractions of proteins
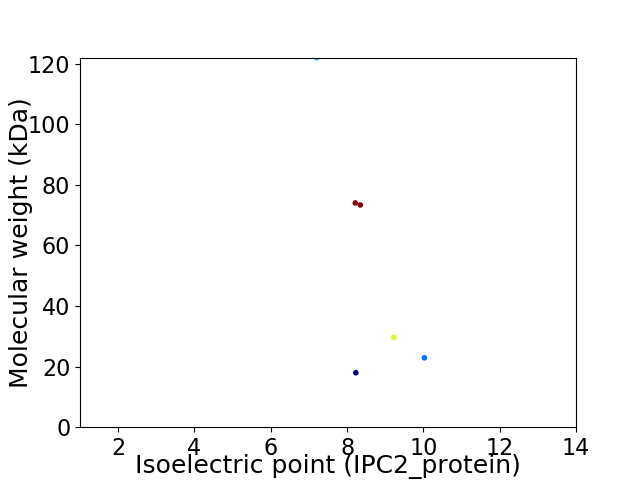
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7EAU5|A0A0A7EAU5_9LUTE Serine protease OS=Sauropus yellowing virus OX=1577997 PE=4 SV=1
MM1 pKa = 7.54AGYY4 pKa = 10.69YY5 pKa = 8.3LTTFLVLLFLFSHH18 pKa = 6.58SSSFPTYY25 pKa = 10.31DD26 pKa = 3.12GRR28 pKa = 11.84VEE30 pKa = 4.56GNFTSTFPMLPPLSNGDD47 pKa = 3.59GGSDD51 pKa = 3.17ISPVSVLMEE60 pKa = 3.94TLYY63 pKa = 11.27VSISDD68 pKa = 4.28EE69 pKa = 4.28PPVAPLTNYY78 pKa = 9.99TYY80 pKa = 11.16GALLGAVSSKK90 pKa = 10.59FVFDD94 pKa = 4.96TIALCNEE101 pKa = 3.93AQEE104 pKa = 4.03IWVTLFRR111 pKa = 11.84SFCDD115 pKa = 3.2NTVNGLKK122 pKa = 9.49WAIEE126 pKa = 3.88KK127 pKa = 10.76LILSLVSIWSWLVYY141 pKa = 9.56QAILCILSCAVNYY154 pKa = 10.71APVLITLVLCGVCTRR169 pKa = 11.84YY170 pKa = 10.48LFLAVIWMYY179 pKa = 10.72GIQVTCLLSFLRR191 pKa = 11.84IISKK195 pKa = 8.07VLRR198 pKa = 11.84FRR200 pKa = 11.84RR201 pKa = 11.84PSGFKK206 pKa = 10.17NEE208 pKa = 3.9KK209 pKa = 9.32SVEE212 pKa = 4.1GFTEE216 pKa = 3.91ITFEE220 pKa = 3.79QSPPGKK226 pKa = 9.95SVLLLEE232 pKa = 4.29EE233 pKa = 4.06RR234 pKa = 11.84RR235 pKa = 11.84NGEE238 pKa = 3.95KK239 pKa = 10.24RR240 pKa = 11.84HH241 pKa = 5.53VGYY244 pKa = 11.09AFAGKK249 pKa = 10.01LYY251 pKa = 10.01NGEE254 pKa = 4.04VGLFTVAHH262 pKa = 6.07NLEE265 pKa = 4.49SCDD268 pKa = 3.85NLWVLSEE275 pKa = 4.1RR276 pKa = 11.84TKK278 pKa = 11.4NSLKK282 pKa = 10.58LKK284 pKa = 9.36QFQPIVLSKK293 pKa = 10.9SADD296 pKa = 3.51LAILRR301 pKa = 11.84GPVNWEE307 pKa = 3.62ALLGCKK313 pKa = 10.12GGIYY317 pKa = 7.07TTSNRR322 pKa = 11.84LAVSGCTWFDD332 pKa = 3.48YY333 pKa = 9.91TEE335 pKa = 3.89SWRR338 pKa = 11.84SHH340 pKa = 4.4SAKK343 pKa = 10.48VVGTDD348 pKa = 3.13GDD350 pKa = 4.1LVRR353 pKa = 11.84VLSNTRR359 pKa = 11.84PGHH362 pKa = 5.96SGTPYY367 pKa = 10.61FNGKK371 pKa = 9.12SCLGLHH377 pKa = 6.08KK378 pKa = 10.29GYY380 pKa = 10.83RR381 pKa = 11.84LAGDD385 pKa = 4.23NNNCMVPIPPIPGYY399 pKa = 6.53TTPDD403 pKa = 3.37YY404 pKa = 10.97KK405 pKa = 11.05FEE407 pKa = 4.16TSAPQGLLFDD417 pKa = 4.52SKK419 pKa = 10.07TIARR423 pKa = 11.84LEE425 pKa = 4.07KK426 pKa = 9.88QAMEE430 pKa = 4.4MVRR433 pKa = 11.84HH434 pKa = 6.24SGGVWDD440 pKa = 5.15FDD442 pKa = 3.94DD443 pKa = 4.46AFAVVSSDD451 pKa = 3.59YY452 pKa = 11.3VGEE455 pKa = 4.32TTLPKK460 pKa = 10.15PDD462 pKa = 3.29VDD464 pKa = 4.09EE465 pKa = 4.6SNKK468 pKa = 9.63IPKK471 pKa = 9.51DD472 pKa = 3.42HH473 pKa = 7.09QSFLRR478 pKa = 11.84DD479 pKa = 3.41YY480 pKa = 9.67GTVLTKK486 pKa = 10.72HH487 pKa = 6.31FGQYY491 pKa = 9.49QSPDD495 pKa = 3.26SAVDD499 pKa = 3.96LNLLFPGVKK508 pKa = 9.55PEE510 pKa = 3.74QAQQKK515 pKa = 8.9KK516 pKa = 9.92SSGNRR521 pKa = 11.84VSRR524 pKa = 11.84AGGKK528 pKa = 7.75NTGIRR533 pKa = 11.84RR534 pKa = 11.84TIHH537 pKa = 4.97PFEE540 pKa = 4.33RR541 pKa = 11.84KK542 pKa = 8.98RR543 pKa = 11.84RR544 pKa = 11.84YY545 pKa = 7.95EE546 pKa = 3.62ANRR549 pKa = 11.84AHH551 pKa = 6.89GCAYY555 pKa = 9.51WADD558 pKa = 3.55QLRR561 pKa = 11.84EE562 pKa = 3.81NRR564 pKa = 11.84RR565 pKa = 11.84TGSRR569 pKa = 11.84QNSEE573 pKa = 4.32FVDD576 pKa = 3.71EE577 pKa = 4.54TTQDD581 pKa = 3.27TRR583 pKa = 11.84SSWRR587 pKa = 11.84KK588 pKa = 8.93EE589 pKa = 3.29EE590 pKa = 3.67ARR592 pKa = 11.84NIRR595 pKa = 11.84EE596 pKa = 4.24HH597 pKa = 6.07FASRR601 pKa = 11.84YY602 pKa = 5.59QWNIPSPTPEE612 pKa = 3.7VEE614 pKa = 4.22GFQACGTFAYY624 pKa = 10.03RR625 pKa = 11.84YY626 pKa = 9.07HH627 pKa = 7.14CKK629 pKa = 9.79QLQEE633 pKa = 4.28SDD635 pKa = 2.62WGRR638 pKa = 11.84FLTGQYY644 pKa = 8.53PALGEE649 pKa = 4.08EE650 pKa = 4.4TRR652 pKa = 11.84GFGWPEE658 pKa = 3.26KK659 pKa = 10.46GAAAEE664 pKa = 4.1LKK666 pKa = 10.29SLKK669 pKa = 10.19LQASRR674 pKa = 11.84WRR676 pKa = 11.84EE677 pKa = 3.44RR678 pKa = 11.84ANRR681 pKa = 11.84AEE683 pKa = 4.23IPTGSARR690 pKa = 11.84QNVIEE695 pKa = 4.13QTVRR699 pKa = 11.84AYY701 pKa = 10.78SAVANTDD708 pKa = 2.95TPLCVRR714 pKa = 11.84SGQLVWSDD722 pKa = 3.98FIEE725 pKa = 4.49DD726 pKa = 4.22FKK728 pKa = 11.44QAVSTLEE735 pKa = 3.7LDD737 pKa = 3.63AGVGIPYY744 pKa = 9.97VEE746 pKa = 4.57FGKK749 pKa = 8.53PTHH752 pKa = 6.66RR753 pKa = 11.84GWIEE757 pKa = 3.79DD758 pKa = 3.81SYY760 pKa = 11.18LAPILAQLTFDD771 pKa = 4.5RR772 pKa = 11.84LQKK775 pKa = 9.96MSEE778 pKa = 4.07VNLAEE783 pKa = 4.14LTSAEE788 pKa = 4.32SLVQLGLCDD797 pKa = 5.01PIRR800 pKa = 11.84TFVKK804 pKa = 10.77GEE806 pKa = 3.68PHH808 pKa = 6.04KK809 pKa = 10.35QAKK812 pKa = 9.74LDD814 pKa = 3.49EE815 pKa = 4.35GRR817 pKa = 11.84YY818 pKa = 8.74RR819 pKa = 11.84LIMSVSLVDD828 pKa = 3.43QMVARR833 pKa = 11.84VLFQQQNKK841 pKa = 9.77RR842 pKa = 11.84EE843 pKa = 3.99IALWRR848 pKa = 11.84AIPSKK853 pKa = 10.43PGFGVSSDD861 pKa = 3.5EE862 pKa = 3.97QVAEE866 pKa = 4.95FISLLAQSVGKK877 pKa = 8.38TPIEE881 pKa = 4.22TVGEE885 pKa = 4.05WKK887 pKa = 10.37KK888 pKa = 11.18YY889 pKa = 10.23IIPTDD894 pKa = 3.4CSGFDD899 pKa = 3.21WSVAEE904 pKa = 4.61WMLDD908 pKa = 2.98DD909 pKa = 5.76DD910 pKa = 4.15MEE912 pKa = 4.33VRR914 pKa = 11.84NRR916 pKa = 11.84LTANNNGLLRR926 pKa = 11.84KK927 pKa = 9.55LRR929 pKa = 11.84AVWLQCIKK937 pKa = 10.82QSVLCLSDD945 pKa = 3.79GSLLAQRR952 pKa = 11.84IPGIQKK958 pKa = 9.39SGSYY962 pKa = 8.27NTSSSNSRR970 pKa = 11.84IRR972 pKa = 11.84VMCALHH978 pKa = 6.98CGAPWAIAMGDD989 pKa = 3.65DD990 pKa = 3.51ALEE993 pKa = 4.51GVGSDD998 pKa = 2.94LALYY1002 pKa = 9.79HH1003 pKa = 6.88KK1004 pKa = 10.6LGLKK1008 pKa = 10.24VEE1010 pKa = 4.27VSPKK1014 pKa = 10.82LEE1016 pKa = 4.26FCSHH1020 pKa = 6.37LFEE1023 pKa = 6.76SEE1025 pKa = 4.09TLALPVNINKK1035 pKa = 8.29MLWKK1039 pKa = 10.33LIYY1042 pKa = 10.0GYY1044 pKa = 11.02NPASGNLEE1052 pKa = 4.13VISNYY1057 pKa = 10.05LAACASVMHH1066 pKa = 6.45EE1067 pKa = 4.3MRR1069 pKa = 11.84HH1070 pKa = 5.89LPDD1073 pKa = 3.79KK1074 pKa = 11.16VDD1076 pKa = 3.86IIKK1079 pKa = 10.15RR1080 pKa = 11.84WLVAPIQPQNN1090 pKa = 3.18
MM1 pKa = 7.54AGYY4 pKa = 10.69YY5 pKa = 8.3LTTFLVLLFLFSHH18 pKa = 6.58SSSFPTYY25 pKa = 10.31DD26 pKa = 3.12GRR28 pKa = 11.84VEE30 pKa = 4.56GNFTSTFPMLPPLSNGDD47 pKa = 3.59GGSDD51 pKa = 3.17ISPVSVLMEE60 pKa = 3.94TLYY63 pKa = 11.27VSISDD68 pKa = 4.28EE69 pKa = 4.28PPVAPLTNYY78 pKa = 9.99TYY80 pKa = 11.16GALLGAVSSKK90 pKa = 10.59FVFDD94 pKa = 4.96TIALCNEE101 pKa = 3.93AQEE104 pKa = 4.03IWVTLFRR111 pKa = 11.84SFCDD115 pKa = 3.2NTVNGLKK122 pKa = 9.49WAIEE126 pKa = 3.88KK127 pKa = 10.76LILSLVSIWSWLVYY141 pKa = 9.56QAILCILSCAVNYY154 pKa = 10.71APVLITLVLCGVCTRR169 pKa = 11.84YY170 pKa = 10.48LFLAVIWMYY179 pKa = 10.72GIQVTCLLSFLRR191 pKa = 11.84IISKK195 pKa = 8.07VLRR198 pKa = 11.84FRR200 pKa = 11.84RR201 pKa = 11.84PSGFKK206 pKa = 10.17NEE208 pKa = 3.9KK209 pKa = 9.32SVEE212 pKa = 4.1GFTEE216 pKa = 3.91ITFEE220 pKa = 3.79QSPPGKK226 pKa = 9.95SVLLLEE232 pKa = 4.29EE233 pKa = 4.06RR234 pKa = 11.84RR235 pKa = 11.84NGEE238 pKa = 3.95KK239 pKa = 10.24RR240 pKa = 11.84HH241 pKa = 5.53VGYY244 pKa = 11.09AFAGKK249 pKa = 10.01LYY251 pKa = 10.01NGEE254 pKa = 4.04VGLFTVAHH262 pKa = 6.07NLEE265 pKa = 4.49SCDD268 pKa = 3.85NLWVLSEE275 pKa = 4.1RR276 pKa = 11.84TKK278 pKa = 11.4NSLKK282 pKa = 10.58LKK284 pKa = 9.36QFQPIVLSKK293 pKa = 10.9SADD296 pKa = 3.51LAILRR301 pKa = 11.84GPVNWEE307 pKa = 3.62ALLGCKK313 pKa = 10.12GGIYY317 pKa = 7.07TTSNRR322 pKa = 11.84LAVSGCTWFDD332 pKa = 3.48YY333 pKa = 9.91TEE335 pKa = 3.89SWRR338 pKa = 11.84SHH340 pKa = 4.4SAKK343 pKa = 10.48VVGTDD348 pKa = 3.13GDD350 pKa = 4.1LVRR353 pKa = 11.84VLSNTRR359 pKa = 11.84PGHH362 pKa = 5.96SGTPYY367 pKa = 10.61FNGKK371 pKa = 9.12SCLGLHH377 pKa = 6.08KK378 pKa = 10.29GYY380 pKa = 10.83RR381 pKa = 11.84LAGDD385 pKa = 4.23NNNCMVPIPPIPGYY399 pKa = 6.53TTPDD403 pKa = 3.37YY404 pKa = 10.97KK405 pKa = 11.05FEE407 pKa = 4.16TSAPQGLLFDD417 pKa = 4.52SKK419 pKa = 10.07TIARR423 pKa = 11.84LEE425 pKa = 4.07KK426 pKa = 9.88QAMEE430 pKa = 4.4MVRR433 pKa = 11.84HH434 pKa = 6.24SGGVWDD440 pKa = 5.15FDD442 pKa = 3.94DD443 pKa = 4.46AFAVVSSDD451 pKa = 3.59YY452 pKa = 11.3VGEE455 pKa = 4.32TTLPKK460 pKa = 10.15PDD462 pKa = 3.29VDD464 pKa = 4.09EE465 pKa = 4.6SNKK468 pKa = 9.63IPKK471 pKa = 9.51DD472 pKa = 3.42HH473 pKa = 7.09QSFLRR478 pKa = 11.84DD479 pKa = 3.41YY480 pKa = 9.67GTVLTKK486 pKa = 10.72HH487 pKa = 6.31FGQYY491 pKa = 9.49QSPDD495 pKa = 3.26SAVDD499 pKa = 3.96LNLLFPGVKK508 pKa = 9.55PEE510 pKa = 3.74QAQQKK515 pKa = 8.9KK516 pKa = 9.92SSGNRR521 pKa = 11.84VSRR524 pKa = 11.84AGGKK528 pKa = 7.75NTGIRR533 pKa = 11.84RR534 pKa = 11.84TIHH537 pKa = 4.97PFEE540 pKa = 4.33RR541 pKa = 11.84KK542 pKa = 8.98RR543 pKa = 11.84RR544 pKa = 11.84YY545 pKa = 7.95EE546 pKa = 3.62ANRR549 pKa = 11.84AHH551 pKa = 6.89GCAYY555 pKa = 9.51WADD558 pKa = 3.55QLRR561 pKa = 11.84EE562 pKa = 3.81NRR564 pKa = 11.84RR565 pKa = 11.84TGSRR569 pKa = 11.84QNSEE573 pKa = 4.32FVDD576 pKa = 3.71EE577 pKa = 4.54TTQDD581 pKa = 3.27TRR583 pKa = 11.84SSWRR587 pKa = 11.84KK588 pKa = 8.93EE589 pKa = 3.29EE590 pKa = 3.67ARR592 pKa = 11.84NIRR595 pKa = 11.84EE596 pKa = 4.24HH597 pKa = 6.07FASRR601 pKa = 11.84YY602 pKa = 5.59QWNIPSPTPEE612 pKa = 3.7VEE614 pKa = 4.22GFQACGTFAYY624 pKa = 10.03RR625 pKa = 11.84YY626 pKa = 9.07HH627 pKa = 7.14CKK629 pKa = 9.79QLQEE633 pKa = 4.28SDD635 pKa = 2.62WGRR638 pKa = 11.84FLTGQYY644 pKa = 8.53PALGEE649 pKa = 4.08EE650 pKa = 4.4TRR652 pKa = 11.84GFGWPEE658 pKa = 3.26KK659 pKa = 10.46GAAAEE664 pKa = 4.1LKK666 pKa = 10.29SLKK669 pKa = 10.19LQASRR674 pKa = 11.84WRR676 pKa = 11.84EE677 pKa = 3.44RR678 pKa = 11.84ANRR681 pKa = 11.84AEE683 pKa = 4.23IPTGSARR690 pKa = 11.84QNVIEE695 pKa = 4.13QTVRR699 pKa = 11.84AYY701 pKa = 10.78SAVANTDD708 pKa = 2.95TPLCVRR714 pKa = 11.84SGQLVWSDD722 pKa = 3.98FIEE725 pKa = 4.49DD726 pKa = 4.22FKK728 pKa = 11.44QAVSTLEE735 pKa = 3.7LDD737 pKa = 3.63AGVGIPYY744 pKa = 9.97VEE746 pKa = 4.57FGKK749 pKa = 8.53PTHH752 pKa = 6.66RR753 pKa = 11.84GWIEE757 pKa = 3.79DD758 pKa = 3.81SYY760 pKa = 11.18LAPILAQLTFDD771 pKa = 4.5RR772 pKa = 11.84LQKK775 pKa = 9.96MSEE778 pKa = 4.07VNLAEE783 pKa = 4.14LTSAEE788 pKa = 4.32SLVQLGLCDD797 pKa = 5.01PIRR800 pKa = 11.84TFVKK804 pKa = 10.77GEE806 pKa = 3.68PHH808 pKa = 6.04KK809 pKa = 10.35QAKK812 pKa = 9.74LDD814 pKa = 3.49EE815 pKa = 4.35GRR817 pKa = 11.84YY818 pKa = 8.74RR819 pKa = 11.84LIMSVSLVDD828 pKa = 3.43QMVARR833 pKa = 11.84VLFQQQNKK841 pKa = 9.77RR842 pKa = 11.84EE843 pKa = 3.99IALWRR848 pKa = 11.84AIPSKK853 pKa = 10.43PGFGVSSDD861 pKa = 3.5EE862 pKa = 3.97QVAEE866 pKa = 4.95FISLLAQSVGKK877 pKa = 8.38TPIEE881 pKa = 4.22TVGEE885 pKa = 4.05WKK887 pKa = 10.37KK888 pKa = 11.18YY889 pKa = 10.23IIPTDD894 pKa = 3.4CSGFDD899 pKa = 3.21WSVAEE904 pKa = 4.61WMLDD908 pKa = 2.98DD909 pKa = 5.76DD910 pKa = 4.15MEE912 pKa = 4.33VRR914 pKa = 11.84NRR916 pKa = 11.84LTANNNGLLRR926 pKa = 11.84KK927 pKa = 9.55LRR929 pKa = 11.84AVWLQCIKK937 pKa = 10.82QSVLCLSDD945 pKa = 3.79GSLLAQRR952 pKa = 11.84IPGIQKK958 pKa = 9.39SGSYY962 pKa = 8.27NTSSSNSRR970 pKa = 11.84IRR972 pKa = 11.84VMCALHH978 pKa = 6.98CGAPWAIAMGDD989 pKa = 3.65DD990 pKa = 3.51ALEE993 pKa = 4.51GVGSDD998 pKa = 2.94LALYY1002 pKa = 9.79HH1003 pKa = 6.88KK1004 pKa = 10.6LGLKK1008 pKa = 10.24VEE1010 pKa = 4.27VSPKK1014 pKa = 10.82LEE1016 pKa = 4.26FCSHH1020 pKa = 6.37LFEE1023 pKa = 6.76SEE1025 pKa = 4.09TLALPVNINKK1035 pKa = 8.29MLWKK1039 pKa = 10.33LIYY1042 pKa = 10.0GYY1044 pKa = 11.02NPASGNLEE1052 pKa = 4.13VISNYY1057 pKa = 10.05LAACASVMHH1066 pKa = 6.45EE1067 pKa = 4.3MRR1069 pKa = 11.84HH1070 pKa = 5.89LPDD1073 pKa = 3.79KK1074 pKa = 11.16VDD1076 pKa = 3.86IIKK1079 pKa = 10.15RR1080 pKa = 11.84WLVAPIQPQNN1090 pKa = 3.18
Molecular weight: 122.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7E9K4|A0A0A7E9K4_9LUTE Readthrough protein OS=Sauropus yellowing virus OX=1577997 PE=3 SV=1
MM1 pKa = 6.62NTAGRR6 pKa = 11.84KK7 pKa = 7.97GRR9 pKa = 11.84RR10 pKa = 11.84GNGKK14 pKa = 9.38RR15 pKa = 11.84RR16 pKa = 11.84FNNVTRR22 pKa = 11.84QQRR25 pKa = 11.84TIQPVVVVTPNGQPRR40 pKa = 11.84GRR42 pKa = 11.84NRR44 pKa = 11.84GRR46 pKa = 11.84RR47 pKa = 11.84NRR49 pKa = 11.84LRR51 pKa = 11.84NRR53 pKa = 11.84GNRR56 pKa = 11.84GRR58 pKa = 11.84TARR61 pKa = 11.84GSVQSEE67 pKa = 4.02TFVFNKK73 pKa = 10.26DD74 pKa = 3.39DD75 pKa = 4.31LKK77 pKa = 11.22GSSHH81 pKa = 5.45GTIKK85 pKa = 10.75FGPNLSEE92 pKa = 4.39SVALSAGVLKK102 pKa = 10.51AYY104 pKa = 10.31HH105 pKa = 6.65EE106 pKa = 4.46YY107 pKa = 10.43KK108 pKa = 10.15IVMVNIRR115 pKa = 11.84FVSEE119 pKa = 4.22SSSTAQGSIAYY130 pKa = 9.28EE131 pKa = 3.94LDD133 pKa = 3.38PHH135 pKa = 7.0CKK137 pKa = 9.85LDD139 pKa = 3.78ALKK142 pKa = 9.62STLRR146 pKa = 11.84KK147 pKa = 9.65FPITKK152 pKa = 10.07GGQATFRR159 pKa = 11.84ASEE162 pKa = 3.93INGEE166 pKa = 4.16KK167 pKa = 9.35WHH169 pKa = 6.92DD170 pKa = 3.53TTVDD174 pKa = 3.49QFRR177 pKa = 11.84LLYY180 pKa = 9.71KK181 pKa = 10.9GNGAASEE188 pKa = 4.24TAGFFQIRR196 pKa = 11.84FTVQLHH202 pKa = 5.06NPKK205 pKa = 10.54
MM1 pKa = 6.62NTAGRR6 pKa = 11.84KK7 pKa = 7.97GRR9 pKa = 11.84RR10 pKa = 11.84GNGKK14 pKa = 9.38RR15 pKa = 11.84RR16 pKa = 11.84FNNVTRR22 pKa = 11.84QQRR25 pKa = 11.84TIQPVVVVTPNGQPRR40 pKa = 11.84GRR42 pKa = 11.84NRR44 pKa = 11.84GRR46 pKa = 11.84RR47 pKa = 11.84NRR49 pKa = 11.84LRR51 pKa = 11.84NRR53 pKa = 11.84GNRR56 pKa = 11.84GRR58 pKa = 11.84TARR61 pKa = 11.84GSVQSEE67 pKa = 4.02TFVFNKK73 pKa = 10.26DD74 pKa = 3.39DD75 pKa = 4.31LKK77 pKa = 11.22GSSHH81 pKa = 5.45GTIKK85 pKa = 10.75FGPNLSEE92 pKa = 4.39SVALSAGVLKK102 pKa = 10.51AYY104 pKa = 10.31HH105 pKa = 6.65EE106 pKa = 4.46YY107 pKa = 10.43KK108 pKa = 10.15IVMVNIRR115 pKa = 11.84FVSEE119 pKa = 4.22SSSTAQGSIAYY130 pKa = 9.28EE131 pKa = 3.94LDD133 pKa = 3.38PHH135 pKa = 7.0CKK137 pKa = 9.85LDD139 pKa = 3.78ALKK142 pKa = 9.62STLRR146 pKa = 11.84KK147 pKa = 9.65FPITKK152 pKa = 10.07GGQATFRR159 pKa = 11.84ASEE162 pKa = 3.93INGEE166 pKa = 4.16KK167 pKa = 9.35WHH169 pKa = 6.92DD170 pKa = 3.53TTVDD174 pKa = 3.49QFRR177 pKa = 11.84LLYY180 pKa = 9.71KK181 pKa = 10.9GNGAASEE188 pKa = 4.24TAGFFQIRR196 pKa = 11.84FTVQLHH202 pKa = 5.06NPKK205 pKa = 10.54
Molecular weight: 22.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3036 |
161 |
1090 |
506.0 |
56.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.192 ± 0.432 | 1.68 ± 0.257 |
4.545 ± 0.502 | 5.567 ± 0.346 |
4.348 ± 0.462 | 7.444 ± 0.46 |
1.91 ± 0.276 | 4.578 ± 0.312 |
5.27 ± 0.492 | 9.519 ± 0.702 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.548 ± 0.258 | 4.348 ± 0.473 |
5.369 ± 0.528 | 3.722 ± 0.419 |
7.016 ± 0.987 | 8.959 ± 0.597 |
6.061 ± 0.465 | 6.719 ± 0.353 |
1.877 ± 0.291 | 3.294 ± 0.29 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
