
Vibrio proteolyticus NBRC 13287
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio; Vibrio proteolyticus
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
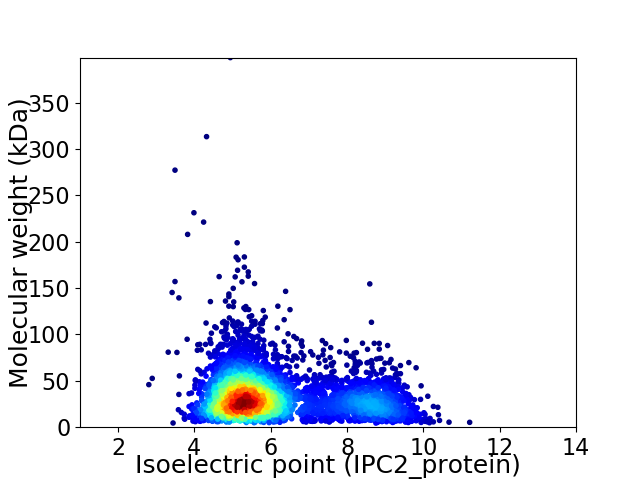
Virtual 2D-PAGE plot for 4347 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U3A5L8|U3A5L8_VIBPR Ancillary SecYEG translocon subunit OS=Vibrio proteolyticus NBRC 13287 OX=1219065 GN=VPR01S_18_00340 PE=3 SV=1
MM1 pKa = 7.63GRR3 pKa = 11.84RR4 pKa = 11.84FKK6 pKa = 11.02NGYY9 pKa = 8.82LRR11 pKa = 11.84AAVVLMLSTLGLVGCNSDD29 pKa = 3.62SDD31 pKa = 5.1DD32 pKa = 4.17DD33 pKa = 4.59VSSQVQALSFNVTNTTDD50 pKa = 3.68DD51 pKa = 3.89TPLSNLEE58 pKa = 3.69VRR60 pKa = 11.84FVFDD64 pKa = 4.84VSNQSQDD71 pKa = 2.97FRR73 pKa = 11.84VSVTDD78 pKa = 3.16VSGRR82 pKa = 11.84EE83 pKa = 3.61VAGYY87 pKa = 10.96VDD89 pKa = 3.77GTAYY93 pKa = 9.99QSNVLLVPDD102 pKa = 4.27SGIVPISISAFNVSSSGDD120 pKa = 3.34SRR122 pKa = 11.84ASLTLVTHH130 pKa = 6.51GKK132 pKa = 10.76GFFSNSQYY140 pKa = 11.03FDD142 pKa = 3.37LTQTSMEE149 pKa = 3.71TSYY152 pKa = 10.29WVEE155 pKa = 3.93VMPKK159 pKa = 10.11SSSDD163 pKa = 3.8DD164 pKa = 3.36VATAYY169 pKa = 9.94AAQIVAMSGSSTASTLTLSTPIQSTTTSGGQILDD203 pKa = 3.74GASASLTIPAGTTLLAEE220 pKa = 5.11DD221 pKa = 5.0GSAFVPVGDD230 pKa = 3.73ITASLMMFSADD241 pKa = 3.46PQGIADD247 pKa = 4.27ADD249 pKa = 3.99NNPLYY254 pKa = 10.38LFPGGLSPQEE264 pKa = 4.03VTGDD268 pKa = 3.83LPADD272 pKa = 3.24IRR274 pKa = 11.84SATGLTFISAGFVAIEE290 pKa = 4.49LEE292 pKa = 4.39DD293 pKa = 3.54EE294 pKa = 5.0AGNQVKK300 pKa = 10.23GFDD303 pKa = 3.59GSGIRR308 pKa = 11.84LTFDD312 pKa = 3.28VPKK315 pKa = 10.27STTNPNTGAPLSLNDD330 pKa = 3.11QTIPIWSYY338 pKa = 11.28TDD340 pKa = 3.31TTGKK344 pKa = 7.78WHH346 pKa = 6.47YY347 pKa = 9.49EE348 pKa = 4.01GEE350 pKa = 4.06ASIVKK355 pKa = 9.83EE356 pKa = 3.96NSDD359 pKa = 3.68TFTLSKK365 pKa = 10.67QITHH369 pKa = 7.01LSYY372 pKa = 11.81YY373 pKa = 10.07NLDD376 pKa = 3.32WYY378 pKa = 10.6DD379 pKa = 3.41QDD381 pKa = 4.42RR382 pKa = 11.84CKK384 pKa = 10.98LDD386 pKa = 3.39INVVDD391 pKa = 5.26QNGEE395 pKa = 4.02PNNQKK400 pKa = 10.18LRR402 pKa = 11.84LSFAKK407 pKa = 10.58AGGGWAYY414 pKa = 9.84KK415 pKa = 10.03PSGWGDD421 pKa = 3.32NPEE424 pKa = 3.87KK425 pKa = 11.14LEE427 pKa = 4.1INRR430 pKa = 11.84VPAFAGHH437 pKa = 6.84FDD439 pKa = 4.39LLDD442 pKa = 3.77SQGNSLLASIEE453 pKa = 3.84VDD455 pKa = 3.53SQVTSVASGDD465 pKa = 3.54TGLDD469 pKa = 3.15LDD471 pKa = 5.0DD472 pKa = 4.66FCFGLSGDD480 pKa = 3.72NTKK483 pKa = 10.66SLKK486 pKa = 9.52ATLNITNPPTIDD498 pKa = 3.15IQPTLKK504 pKa = 10.44LVCPTNTSVAQAVTSGRR521 pKa = 11.84YY522 pKa = 8.37YY523 pKa = 10.82LYY525 pKa = 10.42SGYY528 pKa = 10.49SYY530 pKa = 10.52EE531 pKa = 4.27SSGEE535 pKa = 3.77ISGDD539 pKa = 3.54TLSLTNLLQDD549 pKa = 3.38GLYY552 pKa = 10.37RR553 pKa = 11.84LYY555 pKa = 11.0YY556 pKa = 10.51YY557 pKa = 10.8GGQTWGQVEE566 pKa = 4.74FTASANLDD574 pKa = 4.02EE575 pKa = 4.72IQLMSLEE582 pKa = 4.62LCDD585 pKa = 6.43KK586 pKa = 10.52IDD588 pKa = 3.25QTVNVRR594 pKa = 11.84LVCLDD599 pKa = 3.63DD600 pKa = 3.81QQDD603 pKa = 3.78VTRR606 pKa = 11.84EE607 pKa = 3.73KK608 pKa = 10.57AAPSAYY614 pKa = 8.73YY615 pKa = 9.79WMYY618 pKa = 9.52NQDD621 pKa = 3.18YY622 pKa = 9.82GQYY625 pKa = 10.73LWGQVDD631 pKa = 3.88EE632 pKa = 5.96DD633 pKa = 3.92GTAQEE638 pKa = 4.28SRR640 pKa = 11.84AVDD643 pKa = 3.52TVQYY647 pKa = 10.01EE648 pKa = 4.16GSAYY652 pKa = 10.1VRR654 pKa = 11.84LDD656 pKa = 3.14NQYY659 pKa = 10.42YY660 pKa = 7.82WGEE663 pKa = 4.14RR664 pKa = 11.84QTVTASDD671 pKa = 3.8SQPVVFDD678 pKa = 3.24IALPSDD684 pKa = 3.31HH685 pKa = 6.69TFCSEE690 pKa = 3.73VSTVDD695 pKa = 3.33YY696 pKa = 11.07SGSTLVIADD705 pKa = 3.95TQGMDD710 pKa = 3.0IAATTTLTVTLKK722 pKa = 11.09DD723 pKa = 4.43GDD725 pKa = 4.04NQDD728 pKa = 3.72YY729 pKa = 8.01TQSGGTLEE737 pKa = 4.65LASSPSGLVFTDD749 pKa = 2.85ITDD752 pKa = 3.95NADD755 pKa = 3.05GTYY758 pKa = 10.32SATVSATDD766 pKa = 3.27AGSYY770 pKa = 8.84TISATVGGEE779 pKa = 4.16VLSNTAPLTITAVDD793 pKa = 3.71TSLSAVSPDD802 pKa = 3.11TVAQDD807 pKa = 3.38EE808 pKa = 4.78GSTTNVTLTLRR819 pKa = 11.84QADD822 pKa = 3.82NSVVGHH828 pKa = 6.78GGHH831 pKa = 6.22TVTATGLAIDD841 pKa = 4.74FGLVSVATTDD851 pKa = 4.07HH852 pKa = 7.76DD853 pKa = 4.61DD854 pKa = 3.53GTYY857 pKa = 9.89TLAVTCDD864 pKa = 2.87NGFNGSQDD872 pKa = 2.76INVNVNAVVVDD883 pKa = 3.98TLSVSCNSII892 pKa = 3.23
MM1 pKa = 7.63GRR3 pKa = 11.84RR4 pKa = 11.84FKK6 pKa = 11.02NGYY9 pKa = 8.82LRR11 pKa = 11.84AAVVLMLSTLGLVGCNSDD29 pKa = 3.62SDD31 pKa = 5.1DD32 pKa = 4.17DD33 pKa = 4.59VSSQVQALSFNVTNTTDD50 pKa = 3.68DD51 pKa = 3.89TPLSNLEE58 pKa = 3.69VRR60 pKa = 11.84FVFDD64 pKa = 4.84VSNQSQDD71 pKa = 2.97FRR73 pKa = 11.84VSVTDD78 pKa = 3.16VSGRR82 pKa = 11.84EE83 pKa = 3.61VAGYY87 pKa = 10.96VDD89 pKa = 3.77GTAYY93 pKa = 9.99QSNVLLVPDD102 pKa = 4.27SGIVPISISAFNVSSSGDD120 pKa = 3.34SRR122 pKa = 11.84ASLTLVTHH130 pKa = 6.51GKK132 pKa = 10.76GFFSNSQYY140 pKa = 11.03FDD142 pKa = 3.37LTQTSMEE149 pKa = 3.71TSYY152 pKa = 10.29WVEE155 pKa = 3.93VMPKK159 pKa = 10.11SSSDD163 pKa = 3.8DD164 pKa = 3.36VATAYY169 pKa = 9.94AAQIVAMSGSSTASTLTLSTPIQSTTTSGGQILDD203 pKa = 3.74GASASLTIPAGTTLLAEE220 pKa = 5.11DD221 pKa = 5.0GSAFVPVGDD230 pKa = 3.73ITASLMMFSADD241 pKa = 3.46PQGIADD247 pKa = 4.27ADD249 pKa = 3.99NNPLYY254 pKa = 10.38LFPGGLSPQEE264 pKa = 4.03VTGDD268 pKa = 3.83LPADD272 pKa = 3.24IRR274 pKa = 11.84SATGLTFISAGFVAIEE290 pKa = 4.49LEE292 pKa = 4.39DD293 pKa = 3.54EE294 pKa = 5.0AGNQVKK300 pKa = 10.23GFDD303 pKa = 3.59GSGIRR308 pKa = 11.84LTFDD312 pKa = 3.28VPKK315 pKa = 10.27STTNPNTGAPLSLNDD330 pKa = 3.11QTIPIWSYY338 pKa = 11.28TDD340 pKa = 3.31TTGKK344 pKa = 7.78WHH346 pKa = 6.47YY347 pKa = 9.49EE348 pKa = 4.01GEE350 pKa = 4.06ASIVKK355 pKa = 9.83EE356 pKa = 3.96NSDD359 pKa = 3.68TFTLSKK365 pKa = 10.67QITHH369 pKa = 7.01LSYY372 pKa = 11.81YY373 pKa = 10.07NLDD376 pKa = 3.32WYY378 pKa = 10.6DD379 pKa = 3.41QDD381 pKa = 4.42RR382 pKa = 11.84CKK384 pKa = 10.98LDD386 pKa = 3.39INVVDD391 pKa = 5.26QNGEE395 pKa = 4.02PNNQKK400 pKa = 10.18LRR402 pKa = 11.84LSFAKK407 pKa = 10.58AGGGWAYY414 pKa = 9.84KK415 pKa = 10.03PSGWGDD421 pKa = 3.32NPEE424 pKa = 3.87KK425 pKa = 11.14LEE427 pKa = 4.1INRR430 pKa = 11.84VPAFAGHH437 pKa = 6.84FDD439 pKa = 4.39LLDD442 pKa = 3.77SQGNSLLASIEE453 pKa = 3.84VDD455 pKa = 3.53SQVTSVASGDD465 pKa = 3.54TGLDD469 pKa = 3.15LDD471 pKa = 5.0DD472 pKa = 4.66FCFGLSGDD480 pKa = 3.72NTKK483 pKa = 10.66SLKK486 pKa = 9.52ATLNITNPPTIDD498 pKa = 3.15IQPTLKK504 pKa = 10.44LVCPTNTSVAQAVTSGRR521 pKa = 11.84YY522 pKa = 8.37YY523 pKa = 10.82LYY525 pKa = 10.42SGYY528 pKa = 10.49SYY530 pKa = 10.52EE531 pKa = 4.27SSGEE535 pKa = 3.77ISGDD539 pKa = 3.54TLSLTNLLQDD549 pKa = 3.38GLYY552 pKa = 10.37RR553 pKa = 11.84LYY555 pKa = 11.0YY556 pKa = 10.51YY557 pKa = 10.8GGQTWGQVEE566 pKa = 4.74FTASANLDD574 pKa = 4.02EE575 pKa = 4.72IQLMSLEE582 pKa = 4.62LCDD585 pKa = 6.43KK586 pKa = 10.52IDD588 pKa = 3.25QTVNVRR594 pKa = 11.84LVCLDD599 pKa = 3.63DD600 pKa = 3.81QQDD603 pKa = 3.78VTRR606 pKa = 11.84EE607 pKa = 3.73KK608 pKa = 10.57AAPSAYY614 pKa = 8.73YY615 pKa = 9.79WMYY618 pKa = 9.52NQDD621 pKa = 3.18YY622 pKa = 9.82GQYY625 pKa = 10.73LWGQVDD631 pKa = 3.88EE632 pKa = 5.96DD633 pKa = 3.92GTAQEE638 pKa = 4.28SRR640 pKa = 11.84AVDD643 pKa = 3.52TVQYY647 pKa = 10.01EE648 pKa = 4.16GSAYY652 pKa = 10.1VRR654 pKa = 11.84LDD656 pKa = 3.14NQYY659 pKa = 10.42YY660 pKa = 7.82WGEE663 pKa = 4.14RR664 pKa = 11.84QTVTASDD671 pKa = 3.8SQPVVFDD678 pKa = 3.24IALPSDD684 pKa = 3.31HH685 pKa = 6.69TFCSEE690 pKa = 3.73VSTVDD695 pKa = 3.33YY696 pKa = 11.07SGSTLVIADD705 pKa = 3.95TQGMDD710 pKa = 3.0IAATTTLTVTLKK722 pKa = 11.09DD723 pKa = 4.43GDD725 pKa = 4.04NQDD728 pKa = 3.72YY729 pKa = 8.01TQSGGTLEE737 pKa = 4.65LASSPSGLVFTDD749 pKa = 2.85ITDD752 pKa = 3.95NADD755 pKa = 3.05GTYY758 pKa = 10.32SATVSATDD766 pKa = 3.27AGSYY770 pKa = 8.84TISATVGGEE779 pKa = 4.16VLSNTAPLTITAVDD793 pKa = 3.71TSLSAVSPDD802 pKa = 3.11TVAQDD807 pKa = 3.38EE808 pKa = 4.78GSTTNVTLTLRR819 pKa = 11.84QADD822 pKa = 3.82NSVVGHH828 pKa = 6.78GGHH831 pKa = 6.22TVTATGLAIDD841 pKa = 4.74FGLVSVATTDD851 pKa = 4.07HH852 pKa = 7.76DD853 pKa = 4.61DD854 pKa = 3.53GTYY857 pKa = 9.89TLAVTCDD864 pKa = 2.87NGFNGSQDD872 pKa = 2.76INVNVNAVVVDD883 pKa = 3.98TLSVSCNSII892 pKa = 3.23
Molecular weight: 94.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U2ZMX6|U2ZMX6_VIBPR Cardiolipin synthase A OS=Vibrio proteolyticus NBRC 13287 OX=1219065 GN=cls PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.5RR3 pKa = 11.84TFQPTVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44NGRR28 pKa = 11.84KK29 pKa = 9.39VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSKK44 pKa = 10.84
MM1 pKa = 7.45KK2 pKa = 9.5RR3 pKa = 11.84TFQPTVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44NGRR28 pKa = 11.84KK29 pKa = 9.39VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSKK44 pKa = 10.84
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1389250 |
39 |
3580 |
319.6 |
35.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.099 ± 0.038 | 1.042 ± 0.014 |
5.576 ± 0.039 | 6.078 ± 0.037 |
3.975 ± 0.026 | 6.868 ± 0.038 |
2.377 ± 0.02 | 5.887 ± 0.034 |
4.72 ± 0.033 | 10.579 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.713 ± 0.021 | 3.863 ± 0.024 |
4.052 ± 0.026 | 4.9 ± 0.035 |
4.901 ± 0.031 | 6.443 ± 0.039 |
5.353 ± 0.031 | 7.216 ± 0.031 |
1.302 ± 0.017 | 3.054 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
