
Microbacterium foliorum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria;
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
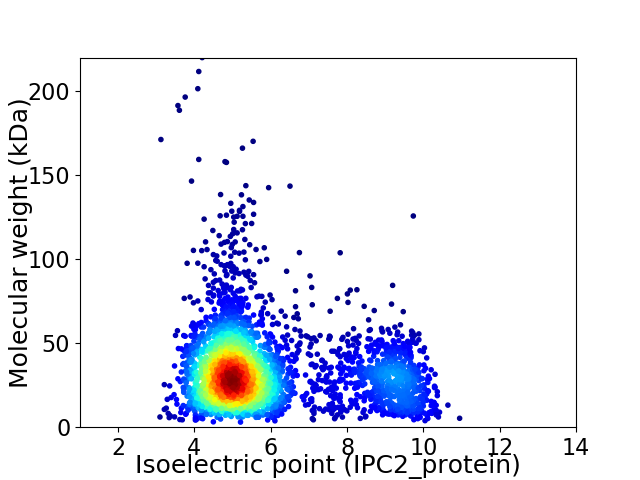
Virtual 2D-PAGE plot for 3324 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F0KRF5|A0A0F0KRF5_9MICO Gamma-aminobutyraldehyde dehydrogenase OS=Microbacterium foliorum OX=104336 GN=prr PE=3 SV=1
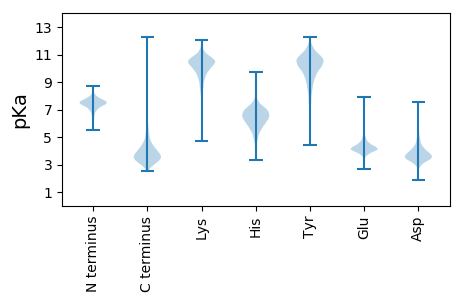
MM1 pKa = 7.63KK2 pKa = 9.0NTKK5 pKa = 9.99RR6 pKa = 11.84RR7 pKa = 11.84WLTGAVATIGAGALALGGAVVPAAADD33 pKa = 3.75DD34 pKa = 4.43AQDD37 pKa = 4.11GPVEE41 pKa = 4.19LQSSASKK48 pKa = 10.84YY49 pKa = 10.78DD50 pKa = 3.43LAAAQSGTSAEE61 pKa = 4.14EE62 pKa = 3.71IKK64 pKa = 10.9NQEE67 pKa = 3.85EE68 pKa = 3.98AGLISVTDD76 pKa = 3.14KK77 pKa = 11.48GFFYY81 pKa = 10.94FNDD84 pKa = 3.88QIHH87 pKa = 6.61EE88 pKa = 4.35SQSLRR93 pKa = 11.84TLALPAAPEE102 pKa = 4.1GAAIPGTPAAGSRR115 pKa = 11.84PGAPVTVYY123 pKa = 11.16LDD125 pKa = 3.62FDD127 pKa = 4.24GEE129 pKa = 4.6TLEE132 pKa = 4.39GTHH135 pKa = 6.62WNTDD139 pKa = 2.87SGIASLPFAPAAAVTDD155 pKa = 4.8RR156 pKa = 11.84AAVWAAVAEE165 pKa = 4.76DD166 pKa = 3.83YY167 pKa = 11.55APFNVNVTTTRR178 pKa = 11.84PSDD181 pKa = 3.43DD182 pKa = 3.85ALYY185 pKa = 8.52KK186 pKa = 10.38TSQDD190 pKa = 3.41DD191 pKa = 3.64NVYY194 pKa = 9.49GSHH197 pKa = 7.25VIITDD202 pKa = 3.34SYY204 pKa = 12.06DD205 pKa = 3.54EE206 pKa = 4.47VLPAAAGNGGIAWLGGTGSDD226 pKa = 4.12YY227 pKa = 10.92LTGALVFTEE236 pKa = 4.5GTGGSPKK243 pKa = 10.48AIAEE247 pKa = 4.26IAAHH251 pKa = 5.95EE252 pKa = 4.57SGHH255 pKa = 6.01NFGLEE260 pKa = 3.65HH261 pKa = 7.3DD262 pKa = 5.17GISGAGGGEE271 pKa = 4.27YY272 pKa = 10.24YY273 pKa = 10.85APEE276 pKa = 5.26DD277 pKa = 4.25GVWGTIMGAAYY288 pKa = 8.91YY289 pKa = 10.57VPVSQWSAGAYY300 pKa = 10.01AGATNDD306 pKa = 3.54QDD308 pKa = 4.42DD309 pKa = 4.49LATITDD315 pKa = 4.16RR316 pKa = 11.84AAGGPLFVNATLPDD330 pKa = 4.11GSPYY334 pKa = 10.05TGNSVCLPAGSTANPSDD351 pKa = 4.24PKK353 pKa = 10.67PGDD356 pKa = 3.27VIFAVGPNGDD366 pKa = 4.66CNPAGPQLTLNFTWTDD382 pKa = 3.4RR383 pKa = 11.84ADD385 pKa = 3.62YY386 pKa = 10.93AADD389 pKa = 3.66TVGNTPEE396 pKa = 4.05TAQTLDD402 pKa = 3.28NAAGTFGAASVIEE415 pKa = 4.5SNSDD419 pKa = 2.78VDD421 pKa = 4.73VFALTTAGGALTANVQVANISPNLDD446 pKa = 3.08AKK448 pKa = 11.0LSVVDD453 pKa = 3.97ASGAVLAEE461 pKa = 4.27NNPEE465 pKa = 4.0STRR468 pKa = 11.84TSDD471 pKa = 3.03TVAAGLNASVTVPNLEE487 pKa = 4.01AGVYY491 pKa = 8.09YY492 pKa = 10.57LAVEE496 pKa = 4.47GVGAGDD502 pKa = 3.84PATATALNANGYY514 pKa = 9.13DD515 pKa = 3.64AYY517 pKa = 10.77GSLGNYY523 pKa = 6.68TLSGAAEE530 pKa = 4.23PFASAPLVILTPEE543 pKa = 4.84DD544 pKa = 3.59GSAVTGGEE552 pKa = 4.24EE553 pKa = 4.04IDD555 pKa = 3.77VTGTATPGATVTLSVGGAAVDD576 pKa = 3.69TVTADD581 pKa = 4.06GAGAWSGNVTVNQYY595 pKa = 11.14DD596 pKa = 3.29ATEE599 pKa = 3.92IVAAQTVGTIAIPGTTSVTVIAPAAPIAAPAITSPTNGATTDD641 pKa = 3.92DD642 pKa = 4.06STPTISGTGVAGATATVTVRR662 pKa = 11.84NATGGEE668 pKa = 4.12LVGQTTVAADD678 pKa = 4.1GAWSLTLTTALANGEE693 pKa = 4.44YY694 pKa = 10.35VVSAVQASAGEE705 pKa = 4.28TSVSSAAVTFTVAAAPPTGNNNNGGGDD732 pKa = 3.76GNLATTGSDD741 pKa = 2.93FDD743 pKa = 3.79AAPFVGGAALLLLIGAGVLVYY764 pKa = 10.58ARR766 pKa = 11.84RR767 pKa = 11.84RR768 pKa = 11.84SSSVDD773 pKa = 2.84AA774 pKa = 5.09
MM1 pKa = 7.63KK2 pKa = 9.0NTKK5 pKa = 9.99RR6 pKa = 11.84RR7 pKa = 11.84WLTGAVATIGAGALALGGAVVPAAADD33 pKa = 3.75DD34 pKa = 4.43AQDD37 pKa = 4.11GPVEE41 pKa = 4.19LQSSASKK48 pKa = 10.84YY49 pKa = 10.78DD50 pKa = 3.43LAAAQSGTSAEE61 pKa = 4.14EE62 pKa = 3.71IKK64 pKa = 10.9NQEE67 pKa = 3.85EE68 pKa = 3.98AGLISVTDD76 pKa = 3.14KK77 pKa = 11.48GFFYY81 pKa = 10.94FNDD84 pKa = 3.88QIHH87 pKa = 6.61EE88 pKa = 4.35SQSLRR93 pKa = 11.84TLALPAAPEE102 pKa = 4.1GAAIPGTPAAGSRR115 pKa = 11.84PGAPVTVYY123 pKa = 11.16LDD125 pKa = 3.62FDD127 pKa = 4.24GEE129 pKa = 4.6TLEE132 pKa = 4.39GTHH135 pKa = 6.62WNTDD139 pKa = 2.87SGIASLPFAPAAAVTDD155 pKa = 4.8RR156 pKa = 11.84AAVWAAVAEE165 pKa = 4.76DD166 pKa = 3.83YY167 pKa = 11.55APFNVNVTTTRR178 pKa = 11.84PSDD181 pKa = 3.43DD182 pKa = 3.85ALYY185 pKa = 8.52KK186 pKa = 10.38TSQDD190 pKa = 3.41DD191 pKa = 3.64NVYY194 pKa = 9.49GSHH197 pKa = 7.25VIITDD202 pKa = 3.34SYY204 pKa = 12.06DD205 pKa = 3.54EE206 pKa = 4.47VLPAAAGNGGIAWLGGTGSDD226 pKa = 4.12YY227 pKa = 10.92LTGALVFTEE236 pKa = 4.5GTGGSPKK243 pKa = 10.48AIAEE247 pKa = 4.26IAAHH251 pKa = 5.95EE252 pKa = 4.57SGHH255 pKa = 6.01NFGLEE260 pKa = 3.65HH261 pKa = 7.3DD262 pKa = 5.17GISGAGGGEE271 pKa = 4.27YY272 pKa = 10.24YY273 pKa = 10.85APEE276 pKa = 5.26DD277 pKa = 4.25GVWGTIMGAAYY288 pKa = 8.91YY289 pKa = 10.57VPVSQWSAGAYY300 pKa = 10.01AGATNDD306 pKa = 3.54QDD308 pKa = 4.42DD309 pKa = 4.49LATITDD315 pKa = 4.16RR316 pKa = 11.84AAGGPLFVNATLPDD330 pKa = 4.11GSPYY334 pKa = 10.05TGNSVCLPAGSTANPSDD351 pKa = 4.24PKK353 pKa = 10.67PGDD356 pKa = 3.27VIFAVGPNGDD366 pKa = 4.66CNPAGPQLTLNFTWTDD382 pKa = 3.4RR383 pKa = 11.84ADD385 pKa = 3.62YY386 pKa = 10.93AADD389 pKa = 3.66TVGNTPEE396 pKa = 4.05TAQTLDD402 pKa = 3.28NAAGTFGAASVIEE415 pKa = 4.5SNSDD419 pKa = 2.78VDD421 pKa = 4.73VFALTTAGGALTANVQVANISPNLDD446 pKa = 3.08AKK448 pKa = 11.0LSVVDD453 pKa = 3.97ASGAVLAEE461 pKa = 4.27NNPEE465 pKa = 4.0STRR468 pKa = 11.84TSDD471 pKa = 3.03TVAAGLNASVTVPNLEE487 pKa = 4.01AGVYY491 pKa = 8.09YY492 pKa = 10.57LAVEE496 pKa = 4.47GVGAGDD502 pKa = 3.84PATATALNANGYY514 pKa = 9.13DD515 pKa = 3.64AYY517 pKa = 10.77GSLGNYY523 pKa = 6.68TLSGAAEE530 pKa = 4.23PFASAPLVILTPEE543 pKa = 4.84DD544 pKa = 3.59GSAVTGGEE552 pKa = 4.24EE553 pKa = 4.04IDD555 pKa = 3.77VTGTATPGATVTLSVGGAAVDD576 pKa = 3.69TVTADD581 pKa = 4.06GAGAWSGNVTVNQYY595 pKa = 11.14DD596 pKa = 3.29ATEE599 pKa = 3.92IVAAQTVGTIAIPGTTSVTVIAPAAPIAAPAITSPTNGATTDD641 pKa = 3.92DD642 pKa = 4.06STPTISGTGVAGATATVTVRR662 pKa = 11.84NATGGEE668 pKa = 4.12LVGQTTVAADD678 pKa = 4.1GAWSLTLTTALANGEE693 pKa = 4.44YY694 pKa = 10.35VVSAVQASAGEE705 pKa = 4.28TSVSSAAVTFTVAAAPPTGNNNNGGGDD732 pKa = 3.76GNLATTGSDD741 pKa = 2.93FDD743 pKa = 3.79AAPFVGGAALLLLIGAGVLVYY764 pKa = 10.58ARR766 pKa = 11.84RR767 pKa = 11.84RR768 pKa = 11.84SSSVDD773 pKa = 2.84AA774 pKa = 5.09
Molecular weight: 76.57 kDa
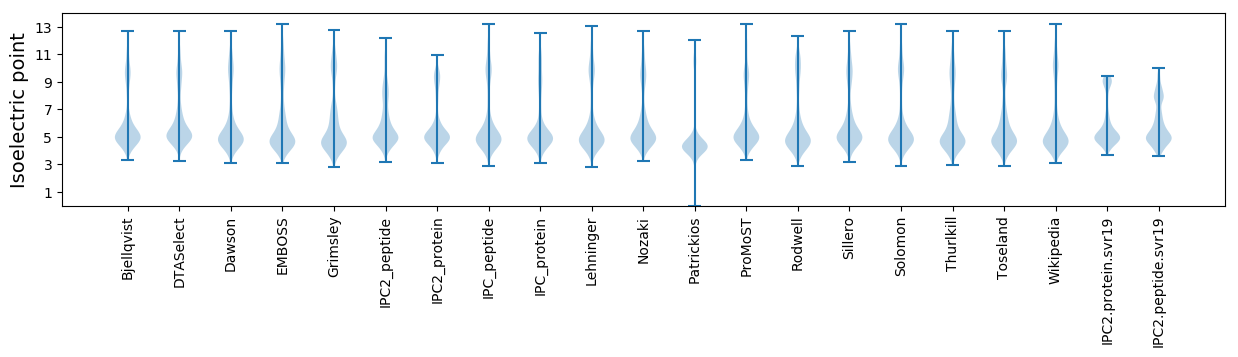
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F0KMS7|A0A0F0KMS7_9MICO Uncharacterized protein OS=Microbacterium foliorum OX=104336 GN=RN50_01637 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.56GRR40 pKa = 11.84TEE42 pKa = 4.14LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.56GRR40 pKa = 11.84TEE42 pKa = 4.14LSAA45 pKa = 4.86
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1078997 |
29 |
2095 |
324.6 |
34.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.397 ± 0.061 | 0.464 ± 0.009 |
6.46 ± 0.039 | 5.609 ± 0.042 |
3.141 ± 0.023 | 8.874 ± 0.036 |
1.938 ± 0.021 | 4.766 ± 0.033 |
1.857 ± 0.031 | 10.059 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.822 ± 0.016 | 1.898 ± 0.025 |
5.265 ± 0.03 | 2.759 ± 0.023 |
7.232 ± 0.051 | 5.912 ± 0.03 |
6.208 ± 0.038 | 8.884 ± 0.039 |
1.526 ± 0.017 | 1.929 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
