
Labeo rohita (Indian major carp) (Cyprinus rohita)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Otomorpha; Ostariophysi; Otophy
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
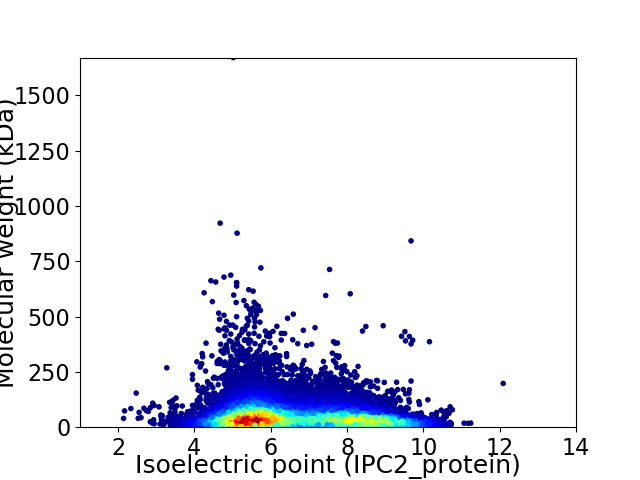
Virtual 2D-PAGE plot for 32379 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
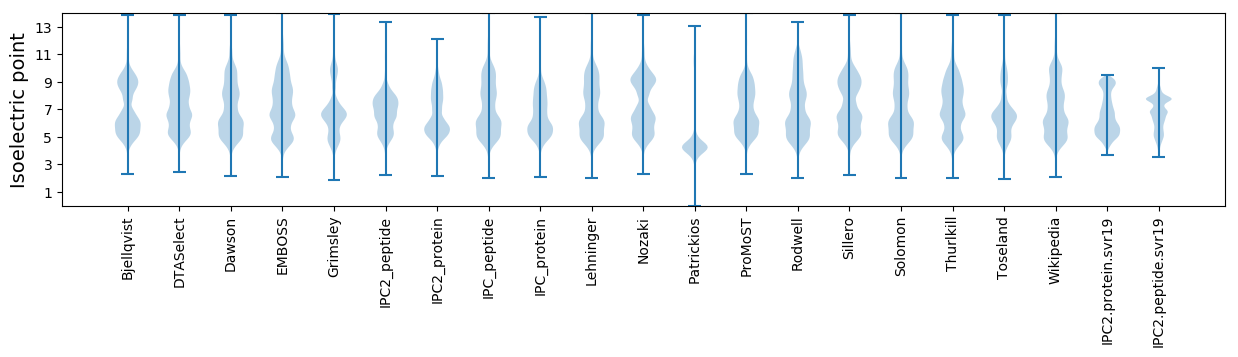
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A498NI06|A0A498NI06_LABRO Butyrophilin subfamily 1 member A1-like isoform X2 OS=Labeo rohita OX=84645 GN=ROHU_004769 PE=3 SV=1
MM1 pKa = 7.31KK2 pKa = 10.49VDD4 pKa = 3.69VDD6 pKa = 3.42WSEE9 pKa = 3.98EE10 pKa = 3.94LSVEE14 pKa = 4.14EE15 pKa = 4.47PVNGLLEE22 pKa = 4.14VDD24 pKa = 4.37NDD26 pKa = 3.53WSEE29 pKa = 4.61DD30 pKa = 3.57PTLEE34 pKa = 4.25GPITSRR40 pKa = 11.84SQVDD44 pKa = 4.21DD45 pKa = 3.8DD46 pKa = 3.98WSDD49 pKa = 3.43KK50 pKa = 10.77VFQDD54 pKa = 4.07LVGDD58 pKa = 4.13LSEE61 pKa = 4.09WSTDD65 pKa = 2.99VLYY68 pKa = 11.02ALDD71 pKa = 3.87GVTDD75 pKa = 4.12KK76 pKa = 11.04FHH78 pKa = 7.81SIQTLVAHH86 pKa = 7.08DD87 pKa = 4.42SSDD90 pKa = 3.92DD91 pKa = 3.69VPEE94 pKa = 4.24ATVGTTSDD102 pKa = 3.49SSDD105 pKa = 3.78DD106 pKa = 3.58VPAATVCATSDD117 pKa = 3.49SSNMSDD123 pKa = 3.55SSNDD127 pKa = 3.34VPQSPVCDD135 pKa = 3.59MSDD138 pKa = 3.24SRR140 pKa = 11.84DD141 pKa = 3.86DD142 pKa = 3.6VPEE145 pKa = 3.94ATVCEE150 pKa = 4.24TSSCSDD156 pKa = 3.84DD157 pKa = 3.65VLQATVCPTSDD168 pKa = 3.95CSDD171 pKa = 4.95GIPSAFLCAVSYY183 pKa = 10.97CSDD186 pKa = 3.44GVHH189 pKa = 6.48PSSYY193 pKa = 11.26SEE195 pKa = 3.92MSYY198 pKa = 11.07SSNDD202 pKa = 3.01GFLEE206 pKa = 3.93ATISGTSSYY215 pKa = 11.23SDD217 pKa = 3.86DD218 pKa = 4.1VPPSSCCDD226 pKa = 3.22MSYY229 pKa = 10.89CSSDD233 pKa = 3.8DD234 pKa = 3.52VLEE237 pKa = 4.39ATACQTEE244 pKa = 4.72DD245 pKa = 3.15TVAPQSNQLQNEE257 pKa = 4.53TKK259 pKa = 10.77VIGQFANITHH269 pKa = 6.65ISIHH273 pKa = 5.49QFMPLWNRR281 pKa = 11.84PSAGKK286 pKa = 9.97RR287 pKa = 11.84RR288 pKa = 11.84IWNGLCPTRR297 pKa = 11.84LDD299 pKa = 4.78DD300 pKa = 4.7GFQVALKK307 pKa = 10.14FVSNRR312 pKa = 11.84NKK314 pKa = 10.44KK315 pKa = 9.83FFLIDD320 pKa = 4.36GYY322 pKa = 10.94SKK324 pKa = 10.38PLPLEE329 pKa = 3.86VALLILASQDD339 pKa = 3.35PPVPEE344 pKa = 3.92IVEE347 pKa = 3.99LLDD350 pKa = 3.32WDD352 pKa = 4.4VEE354 pKa = 4.07PDD356 pKa = 3.28HH357 pKa = 6.94YY358 pKa = 11.17ILVLEE363 pKa = 4.51RR364 pKa = 11.84PMSFVEE370 pKa = 4.36LNWFVLRR377 pKa = 11.84QIMSLEE383 pKa = 3.86EE384 pKa = 4.09DD385 pKa = 3.83VAGSSCGRR393 pKa = 11.84QYY395 pKa = 11.32VQLSRR400 pKa = 11.84AADD403 pKa = 3.38VEE405 pKa = 4.63YY406 pKa = 9.44CTGILRR412 pKa = 11.84WKK414 pKa = 9.44TSS416 pKa = 2.83
MM1 pKa = 7.31KK2 pKa = 10.49VDD4 pKa = 3.69VDD6 pKa = 3.42WSEE9 pKa = 3.98EE10 pKa = 3.94LSVEE14 pKa = 4.14EE15 pKa = 4.47PVNGLLEE22 pKa = 4.14VDD24 pKa = 4.37NDD26 pKa = 3.53WSEE29 pKa = 4.61DD30 pKa = 3.57PTLEE34 pKa = 4.25GPITSRR40 pKa = 11.84SQVDD44 pKa = 4.21DD45 pKa = 3.8DD46 pKa = 3.98WSDD49 pKa = 3.43KK50 pKa = 10.77VFQDD54 pKa = 4.07LVGDD58 pKa = 4.13LSEE61 pKa = 4.09WSTDD65 pKa = 2.99VLYY68 pKa = 11.02ALDD71 pKa = 3.87GVTDD75 pKa = 4.12KK76 pKa = 11.04FHH78 pKa = 7.81SIQTLVAHH86 pKa = 7.08DD87 pKa = 4.42SSDD90 pKa = 3.92DD91 pKa = 3.69VPEE94 pKa = 4.24ATVGTTSDD102 pKa = 3.49SSDD105 pKa = 3.78DD106 pKa = 3.58VPAATVCATSDD117 pKa = 3.49SSNMSDD123 pKa = 3.55SSNDD127 pKa = 3.34VPQSPVCDD135 pKa = 3.59MSDD138 pKa = 3.24SRR140 pKa = 11.84DD141 pKa = 3.86DD142 pKa = 3.6VPEE145 pKa = 3.94ATVCEE150 pKa = 4.24TSSCSDD156 pKa = 3.84DD157 pKa = 3.65VLQATVCPTSDD168 pKa = 3.95CSDD171 pKa = 4.95GIPSAFLCAVSYY183 pKa = 10.97CSDD186 pKa = 3.44GVHH189 pKa = 6.48PSSYY193 pKa = 11.26SEE195 pKa = 3.92MSYY198 pKa = 11.07SSNDD202 pKa = 3.01GFLEE206 pKa = 3.93ATISGTSSYY215 pKa = 11.23SDD217 pKa = 3.86DD218 pKa = 4.1VPPSSCCDD226 pKa = 3.22MSYY229 pKa = 10.89CSSDD233 pKa = 3.8DD234 pKa = 3.52VLEE237 pKa = 4.39ATACQTEE244 pKa = 4.72DD245 pKa = 3.15TVAPQSNQLQNEE257 pKa = 4.53TKK259 pKa = 10.77VIGQFANITHH269 pKa = 6.65ISIHH273 pKa = 5.49QFMPLWNRR281 pKa = 11.84PSAGKK286 pKa = 9.97RR287 pKa = 11.84RR288 pKa = 11.84IWNGLCPTRR297 pKa = 11.84LDD299 pKa = 4.78DD300 pKa = 4.7GFQVALKK307 pKa = 10.14FVSNRR312 pKa = 11.84NKK314 pKa = 10.44KK315 pKa = 9.83FFLIDD320 pKa = 4.36GYY322 pKa = 10.94SKK324 pKa = 10.38PLPLEE329 pKa = 3.86VALLILASQDD339 pKa = 3.35PPVPEE344 pKa = 3.92IVEE347 pKa = 3.99LLDD350 pKa = 3.32WDD352 pKa = 4.4VEE354 pKa = 4.07PDD356 pKa = 3.28HH357 pKa = 6.94YY358 pKa = 11.17ILVLEE363 pKa = 4.51RR364 pKa = 11.84PMSFVEE370 pKa = 4.36LNWFVLRR377 pKa = 11.84QIMSLEE383 pKa = 3.86EE384 pKa = 4.09DD385 pKa = 3.83VAGSSCGRR393 pKa = 11.84QYY395 pKa = 11.32VQLSRR400 pKa = 11.84AADD403 pKa = 3.38VEE405 pKa = 4.63YY406 pKa = 9.44CTGILRR412 pKa = 11.84WKK414 pKa = 9.44TSS416 pKa = 2.83
Molecular weight: 45.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A498MHH9|A0A498MHH9_LABRO Tyrosine-protein kinase 223-like OS=Labeo rohita OX=84645 GN=ROHU_035416 PE=4 SV=1
MM1 pKa = 7.59LGFTRR6 pKa = 11.84LQTKK10 pKa = 10.26LGFIRR15 pKa = 11.84LQTKK19 pKa = 10.4LGFTRR24 pKa = 11.84LQTKK28 pKa = 10.42LGFTRR33 pKa = 11.84LQTKK37 pKa = 10.42LGFTRR42 pKa = 11.84LQNMLGFTRR51 pKa = 11.84LQTKK55 pKa = 10.26LGFIRR60 pKa = 11.84LQTKK64 pKa = 10.4LGFTRR69 pKa = 11.84LQTKK73 pKa = 10.42LGFTRR78 pKa = 11.84LQTKK82 pKa = 10.2LGFIRR87 pKa = 11.84LPTRR91 pKa = 11.84LGFTRR96 pKa = 11.84LQTRR100 pKa = 11.84MGFTRR105 pKa = 11.84LQTKK109 pKa = 10.2LGFIRR114 pKa = 11.84LPTRR118 pKa = 11.84LGFTRR123 pKa = 11.84LQTRR127 pKa = 11.84MGFTRR132 pKa = 11.84LQTRR136 pKa = 11.84MGFTRR141 pKa = 11.84LQTRR145 pKa = 11.84LGFIKK150 pKa = 9.69ITDD153 pKa = 3.61QAGLHH158 pKa = 5.83
MM1 pKa = 7.59LGFTRR6 pKa = 11.84LQTKK10 pKa = 10.26LGFIRR15 pKa = 11.84LQTKK19 pKa = 10.4LGFTRR24 pKa = 11.84LQTKK28 pKa = 10.42LGFTRR33 pKa = 11.84LQTKK37 pKa = 10.42LGFTRR42 pKa = 11.84LQNMLGFTRR51 pKa = 11.84LQTKK55 pKa = 10.26LGFIRR60 pKa = 11.84LQTKK64 pKa = 10.4LGFTRR69 pKa = 11.84LQTKK73 pKa = 10.42LGFTRR78 pKa = 11.84LQTKK82 pKa = 10.2LGFIRR87 pKa = 11.84LPTRR91 pKa = 11.84LGFTRR96 pKa = 11.84LQTRR100 pKa = 11.84MGFTRR105 pKa = 11.84LQTKK109 pKa = 10.2LGFIRR114 pKa = 11.84LPTRR118 pKa = 11.84LGFTRR123 pKa = 11.84LQTRR127 pKa = 11.84MGFTRR132 pKa = 11.84LQTRR136 pKa = 11.84MGFTRR141 pKa = 11.84LQTRR145 pKa = 11.84LGFIKK150 pKa = 9.69ITDD153 pKa = 3.61QAGLHH158 pKa = 5.83
Molecular weight: 18.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
16615359 |
54 |
13795 |
513.2 |
57.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.465 ± 0.016 | 2.202 ± 0.012 |
5.334 ± 0.013 | 6.977 ± 0.025 |
3.662 ± 0.011 | 5.978 ± 0.019 |
2.537 ± 0.006 | 4.681 ± 0.013 |
5.843 ± 0.019 | 9.341 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.374 ± 0.007 | 4.067 ± 0.017 |
5.412 ± 0.019 | 4.648 ± 0.018 |
5.627 ± 0.016 | 8.481 ± 0.019 |
5.792 ± 0.018 | 6.556 ± 0.018 |
1.177 ± 0.005 | 2.844 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
