
Klebsiella phage ST11-VIM1phi8.2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
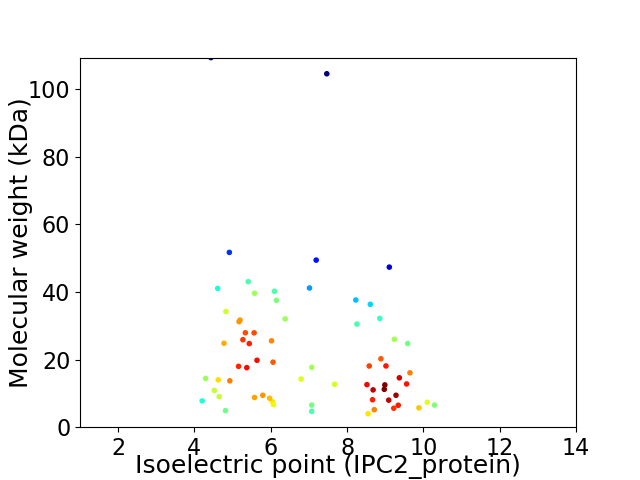
Virtual 2D-PAGE plot for 68 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A482J747|A0A482J747_9CAUD ParA-like protein OS=Klebsiella phage ST11-VIM1phi8.2 OX=2555911 PE=4 SV=1
MM1 pKa = 7.94TIRR4 pKa = 11.84FYY6 pKa = 10.61PSRR9 pKa = 11.84LPGEE13 pKa = 4.03PLEE16 pKa = 4.29THH18 pKa = 5.09EE19 pKa = 5.4HH20 pKa = 5.91GVTSIRR26 pKa = 11.84KK27 pKa = 7.67WLVANVEE34 pKa = 4.39GYY36 pKa = 9.82EE37 pKa = 4.18DD38 pKa = 4.67RR39 pKa = 11.84DD40 pKa = 3.91VPPLTVEE47 pKa = 4.17VEE49 pKa = 4.27GLLIPPGEE57 pKa = 3.92WAKK60 pKa = 10.88CVIRR64 pKa = 11.84PDD66 pKa = 3.34SDD68 pKa = 3.21VRR70 pKa = 11.84LYY72 pKa = 9.71PVPFGLEE79 pKa = 3.51AATIAWIGVGISVAAAAYY97 pKa = 10.08SYY99 pKa = 11.76FMMSNIDD106 pKa = 3.32TGGYY110 pKa = 7.72TSSTGRR116 pKa = 11.84SLDD119 pKa = 3.85LNPAKK124 pKa = 10.84ANTAKK129 pKa = 10.79LGDD132 pKa = 4.4AIRR135 pKa = 11.84EE136 pKa = 3.98VFGRR140 pKa = 11.84VRR142 pKa = 11.84IYY144 pKa = 10.21PDD146 pKa = 3.47YY147 pKa = 10.72VVQPVTRR154 pKa = 11.84FDD156 pKa = 3.6AADD159 pKa = 3.58PTKK162 pKa = 10.46MRR164 pKa = 11.84VQMLLCLGVGDD175 pKa = 5.83LIYY178 pKa = 10.63TNGDD182 pKa = 2.7IRR184 pKa = 11.84VGSTPASTLPGFSSTHH200 pKa = 5.3YY201 pKa = 10.41PPGADD206 pKa = 2.83VSGDD210 pKa = 3.34EE211 pKa = 5.5RR212 pKa = 11.84SEE214 pKa = 3.65NWVNSTEE221 pKa = 4.14VGGTSSGTGLDD232 pKa = 3.72MAQTSPDD239 pKa = 3.77ADD241 pKa = 4.73DD242 pKa = 5.4IIADD246 pKa = 3.75SMTVSGSSVTFTGLDD261 pKa = 3.42TDD263 pKa = 5.62DD264 pKa = 6.56DD265 pKa = 4.87DD266 pKa = 7.47DD267 pKa = 5.91NDD269 pKa = 4.01EE270 pKa = 4.96NDD272 pKa = 3.52NALPPSWVAGAVVEE286 pKa = 4.79LKK288 pKa = 10.93APANYY293 pKa = 10.15QITTAAGYY301 pKa = 9.91SVIASPLLTEE311 pKa = 4.5IAPVVGMPVTLGFNSVDD328 pKa = 3.22YY329 pKa = 11.24DD330 pKa = 4.15LFIASYY336 pKa = 9.69TPGQAAVPGTGGSAAKK352 pKa = 9.71VQASAAPTTYY362 pKa = 10.86DD363 pKa = 3.49FSTSSSTFTITWKK376 pKa = 10.61GVTYY380 pKa = 9.93PVSLVANYY388 pKa = 10.54VSMSGLLAAITEE400 pKa = 4.33GLTGSGLVAQDD411 pKa = 3.17NGGTVLITEE420 pKa = 4.44SASPFAGGAITSSSLPAAVFGDD442 pKa = 3.66APVYY446 pKa = 10.1TSGTASTGGSPAVTANVTLAYY467 pKa = 10.73NSATGTAFSGMPEE480 pKa = 4.26GVQRR484 pKa = 11.84LSLAHH489 pKa = 7.04RR490 pKa = 11.84GNEE493 pKa = 3.85YY494 pKa = 10.51RR495 pKa = 11.84IVSADD500 pKa = 3.01GTTATVARR508 pKa = 11.84LVSGAVDD515 pKa = 3.71EE516 pKa = 4.59SWPGFTARR524 pKa = 11.84TMIDD528 pKa = 3.46YY529 pKa = 8.18EE530 pKa = 4.29ATGLNDD536 pKa = 3.52TLSWLGPFLVCPEE549 pKa = 4.35NEE551 pKa = 4.06VVDD554 pKa = 3.95AFEE557 pKa = 5.04VNFSFPNGICGFDD570 pKa = 3.23SKK572 pKa = 11.14GKK574 pKa = 9.73KK575 pKa = 9.55RR576 pKa = 11.84IRR578 pKa = 11.84HH579 pKa = 4.88VEE581 pKa = 3.4WEE583 pKa = 3.79IQYY586 pKa = 10.44RR587 pKa = 11.84VYY589 pKa = 11.08GSGSGWVSHH598 pKa = 5.62QGEE601 pKa = 4.38YY602 pKa = 10.89ALKK605 pKa = 9.96NVNGLGFTEE614 pKa = 5.93RR615 pKa = 11.84ITLSSPGLVEE625 pKa = 3.87VRR627 pKa = 11.84CRR629 pKa = 11.84RR630 pKa = 11.84RR631 pKa = 11.84NEE633 pKa = 3.79QGSNNARR640 pKa = 11.84DD641 pKa = 3.61SMYY644 pKa = 9.71WQALRR649 pKa = 11.84GRR651 pKa = 11.84LLTRR655 pKa = 11.84PSSYY659 pKa = 10.6PGVSLMAVTVEE670 pKa = 4.3TGGKK674 pKa = 9.22LAAQSDD680 pKa = 3.73RR681 pKa = 11.84RR682 pKa = 11.84VNVVATRR689 pKa = 11.84AYY691 pKa = 10.67DD692 pKa = 3.19SGTARR697 pKa = 11.84TISGALLHH705 pKa = 6.04VANSLGLEE713 pKa = 4.01MDD715 pKa = 3.24VDD717 pKa = 4.66TINALEE723 pKa = 4.09SAYY726 pKa = 6.94WTPRR730 pKa = 11.84GEE732 pKa = 4.31NFDD735 pKa = 3.99FATGDD740 pKa = 3.79SISALEE746 pKa = 4.11MLQKK750 pKa = 10.08IANAGKK756 pKa = 10.28SRR758 pKa = 11.84FLLSDD763 pKa = 3.51GLATVNRR770 pKa = 11.84EE771 pKa = 4.13GIKK774 pKa = 10.0PWTGVITPHH783 pKa = 6.1EE784 pKa = 4.38MVEE787 pKa = 4.11EE788 pKa = 4.13LQSGFTVPSDD798 pKa = 3.69DD799 pKa = 5.46DD800 pKa = 4.43FDD802 pKa = 5.26GVDD805 pKa = 3.17VTYY808 pKa = 10.92INGTTWAEE816 pKa = 3.93EE817 pKa = 4.16TVKK820 pKa = 10.51CRR822 pKa = 11.84TPDD825 pKa = 2.88NPTPVKK831 pKa = 10.28IEE833 pKa = 3.85NYY835 pKa = 10.1KK836 pKa = 10.56LDD838 pKa = 3.65GVLTQDD844 pKa = 2.92HH845 pKa = 7.35AYY847 pKa = 9.43QIGMRR852 pKa = 11.84RR853 pKa = 11.84LMKK856 pKa = 10.65YY857 pKa = 10.14LLQRR861 pKa = 11.84VTFQTTTEE869 pKa = 4.14LDD871 pKa = 3.51ALCYY875 pKa = 8.91NTGDD879 pKa = 4.35RR880 pKa = 11.84IVLTDD885 pKa = 5.8DD886 pKa = 3.24IPGNNTISCLVEE898 pKa = 3.99AMTTAGGVTTFTVTEE913 pKa = 4.01PLDD916 pKa = 3.4WSFEE920 pKa = 4.07NPRR923 pKa = 11.84ALIRR927 pKa = 11.84YY928 pKa = 8.54QDD930 pKa = 3.71GSASGLMVASRR941 pKa = 11.84VGDD944 pKa = 3.66FQLSVPHH951 pKa = 7.11LSEE954 pKa = 5.04FDD956 pKa = 4.36DD957 pKa = 3.86PTKK960 pKa = 10.91VDD962 pKa = 4.16LSSATIEE969 pKa = 4.29PIRR972 pKa = 11.84LVFCGSTRR980 pKa = 11.84HH981 pKa = 5.68VYY983 pKa = 10.15DD984 pKa = 5.57AIVEE988 pKa = 4.53EE989 pKa = 4.87IAPQSDD995 pKa = 3.74GTCQVTAKK1003 pKa = 10.17EE1004 pKa = 4.15YY1005 pKa = 11.23LEE1007 pKa = 5.3SFYY1010 pKa = 11.0QYY1012 pKa = 11.67DD1013 pKa = 3.65DD1014 pKa = 3.0ATYY1017 pKa = 10.17PGDD1020 pKa = 3.5AAA1022 pKa = 4.93
MM1 pKa = 7.94TIRR4 pKa = 11.84FYY6 pKa = 10.61PSRR9 pKa = 11.84LPGEE13 pKa = 4.03PLEE16 pKa = 4.29THH18 pKa = 5.09EE19 pKa = 5.4HH20 pKa = 5.91GVTSIRR26 pKa = 11.84KK27 pKa = 7.67WLVANVEE34 pKa = 4.39GYY36 pKa = 9.82EE37 pKa = 4.18DD38 pKa = 4.67RR39 pKa = 11.84DD40 pKa = 3.91VPPLTVEE47 pKa = 4.17VEE49 pKa = 4.27GLLIPPGEE57 pKa = 3.92WAKK60 pKa = 10.88CVIRR64 pKa = 11.84PDD66 pKa = 3.34SDD68 pKa = 3.21VRR70 pKa = 11.84LYY72 pKa = 9.71PVPFGLEE79 pKa = 3.51AATIAWIGVGISVAAAAYY97 pKa = 10.08SYY99 pKa = 11.76FMMSNIDD106 pKa = 3.32TGGYY110 pKa = 7.72TSSTGRR116 pKa = 11.84SLDD119 pKa = 3.85LNPAKK124 pKa = 10.84ANTAKK129 pKa = 10.79LGDD132 pKa = 4.4AIRR135 pKa = 11.84EE136 pKa = 3.98VFGRR140 pKa = 11.84VRR142 pKa = 11.84IYY144 pKa = 10.21PDD146 pKa = 3.47YY147 pKa = 10.72VVQPVTRR154 pKa = 11.84FDD156 pKa = 3.6AADD159 pKa = 3.58PTKK162 pKa = 10.46MRR164 pKa = 11.84VQMLLCLGVGDD175 pKa = 5.83LIYY178 pKa = 10.63TNGDD182 pKa = 2.7IRR184 pKa = 11.84VGSTPASTLPGFSSTHH200 pKa = 5.3YY201 pKa = 10.41PPGADD206 pKa = 2.83VSGDD210 pKa = 3.34EE211 pKa = 5.5RR212 pKa = 11.84SEE214 pKa = 3.65NWVNSTEE221 pKa = 4.14VGGTSSGTGLDD232 pKa = 3.72MAQTSPDD239 pKa = 3.77ADD241 pKa = 4.73DD242 pKa = 5.4IIADD246 pKa = 3.75SMTVSGSSVTFTGLDD261 pKa = 3.42TDD263 pKa = 5.62DD264 pKa = 6.56DD265 pKa = 4.87DD266 pKa = 7.47DD267 pKa = 5.91NDD269 pKa = 4.01EE270 pKa = 4.96NDD272 pKa = 3.52NALPPSWVAGAVVEE286 pKa = 4.79LKK288 pKa = 10.93APANYY293 pKa = 10.15QITTAAGYY301 pKa = 9.91SVIASPLLTEE311 pKa = 4.5IAPVVGMPVTLGFNSVDD328 pKa = 3.22YY329 pKa = 11.24DD330 pKa = 4.15LFIASYY336 pKa = 9.69TPGQAAVPGTGGSAAKK352 pKa = 9.71VQASAAPTTYY362 pKa = 10.86DD363 pKa = 3.49FSTSSSTFTITWKK376 pKa = 10.61GVTYY380 pKa = 9.93PVSLVANYY388 pKa = 10.54VSMSGLLAAITEE400 pKa = 4.33GLTGSGLVAQDD411 pKa = 3.17NGGTVLITEE420 pKa = 4.44SASPFAGGAITSSSLPAAVFGDD442 pKa = 3.66APVYY446 pKa = 10.1TSGTASTGGSPAVTANVTLAYY467 pKa = 10.73NSATGTAFSGMPEE480 pKa = 4.26GVQRR484 pKa = 11.84LSLAHH489 pKa = 7.04RR490 pKa = 11.84GNEE493 pKa = 3.85YY494 pKa = 10.51RR495 pKa = 11.84IVSADD500 pKa = 3.01GTTATVARR508 pKa = 11.84LVSGAVDD515 pKa = 3.71EE516 pKa = 4.59SWPGFTARR524 pKa = 11.84TMIDD528 pKa = 3.46YY529 pKa = 8.18EE530 pKa = 4.29ATGLNDD536 pKa = 3.52TLSWLGPFLVCPEE549 pKa = 4.35NEE551 pKa = 4.06VVDD554 pKa = 3.95AFEE557 pKa = 5.04VNFSFPNGICGFDD570 pKa = 3.23SKK572 pKa = 11.14GKK574 pKa = 9.73KK575 pKa = 9.55RR576 pKa = 11.84IRR578 pKa = 11.84HH579 pKa = 4.88VEE581 pKa = 3.4WEE583 pKa = 3.79IQYY586 pKa = 10.44RR587 pKa = 11.84VYY589 pKa = 11.08GSGSGWVSHH598 pKa = 5.62QGEE601 pKa = 4.38YY602 pKa = 10.89ALKK605 pKa = 9.96NVNGLGFTEE614 pKa = 5.93RR615 pKa = 11.84ITLSSPGLVEE625 pKa = 3.87VRR627 pKa = 11.84CRR629 pKa = 11.84RR630 pKa = 11.84RR631 pKa = 11.84NEE633 pKa = 3.79QGSNNARR640 pKa = 11.84DD641 pKa = 3.61SMYY644 pKa = 9.71WQALRR649 pKa = 11.84GRR651 pKa = 11.84LLTRR655 pKa = 11.84PSSYY659 pKa = 10.6PGVSLMAVTVEE670 pKa = 4.3TGGKK674 pKa = 9.22LAAQSDD680 pKa = 3.73RR681 pKa = 11.84RR682 pKa = 11.84VNVVATRR689 pKa = 11.84AYY691 pKa = 10.67DD692 pKa = 3.19SGTARR697 pKa = 11.84TISGALLHH705 pKa = 6.04VANSLGLEE713 pKa = 4.01MDD715 pKa = 3.24VDD717 pKa = 4.66TINALEE723 pKa = 4.09SAYY726 pKa = 6.94WTPRR730 pKa = 11.84GEE732 pKa = 4.31NFDD735 pKa = 3.99FATGDD740 pKa = 3.79SISALEE746 pKa = 4.11MLQKK750 pKa = 10.08IANAGKK756 pKa = 10.28SRR758 pKa = 11.84FLLSDD763 pKa = 3.51GLATVNRR770 pKa = 11.84EE771 pKa = 4.13GIKK774 pKa = 10.0PWTGVITPHH783 pKa = 6.1EE784 pKa = 4.38MVEE787 pKa = 4.11EE788 pKa = 4.13LQSGFTVPSDD798 pKa = 3.69DD799 pKa = 5.46DD800 pKa = 4.43FDD802 pKa = 5.26GVDD805 pKa = 3.17VTYY808 pKa = 10.92INGTTWAEE816 pKa = 3.93EE817 pKa = 4.16TVKK820 pKa = 10.51CRR822 pKa = 11.84TPDD825 pKa = 2.88NPTPVKK831 pKa = 10.28IEE833 pKa = 3.85NYY835 pKa = 10.1KK836 pKa = 10.56LDD838 pKa = 3.65GVLTQDD844 pKa = 2.92HH845 pKa = 7.35AYY847 pKa = 9.43QIGMRR852 pKa = 11.84RR853 pKa = 11.84LMKK856 pKa = 10.65YY857 pKa = 10.14LLQRR861 pKa = 11.84VTFQTTTEE869 pKa = 4.14LDD871 pKa = 3.51ALCYY875 pKa = 8.91NTGDD879 pKa = 4.35RR880 pKa = 11.84IVLTDD885 pKa = 5.8DD886 pKa = 3.24IPGNNTISCLVEE898 pKa = 3.99AMTTAGGVTTFTVTEE913 pKa = 4.01PLDD916 pKa = 3.4WSFEE920 pKa = 4.07NPRR923 pKa = 11.84ALIRR927 pKa = 11.84YY928 pKa = 8.54QDD930 pKa = 3.71GSASGLMVASRR941 pKa = 11.84VGDD944 pKa = 3.66FQLSVPHH951 pKa = 7.11LSEE954 pKa = 5.04FDD956 pKa = 4.36DD957 pKa = 3.86PTKK960 pKa = 10.91VDD962 pKa = 4.16LSSATIEE969 pKa = 4.29PIRR972 pKa = 11.84LVFCGSTRR980 pKa = 11.84HH981 pKa = 5.68VYY983 pKa = 10.15DD984 pKa = 5.57AIVEE988 pKa = 4.53EE989 pKa = 4.87IAPQSDD995 pKa = 3.74GTCQVTAKK1003 pKa = 10.17EE1004 pKa = 4.15YY1005 pKa = 11.23LEE1007 pKa = 5.3SFYY1010 pKa = 11.0QYY1012 pKa = 11.67DD1013 pKa = 3.65DD1014 pKa = 3.0ATYY1017 pKa = 10.17PGDD1020 pKa = 3.5AAA1022 pKa = 4.93
Molecular weight: 109.25 kDa
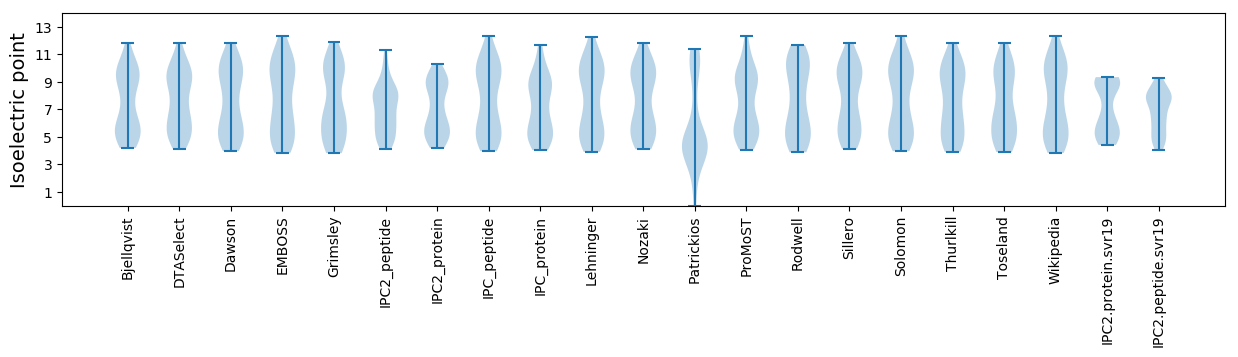
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A482J393|A0A482J393_9CAUD CII family protein OS=Klebsiella phage ST11-VIM1phi8.2 OX=2555911 PE=4 SV=1
MM1 pKa = 7.89RR2 pKa = 11.84KK3 pKa = 8.26PARR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.32CAHH11 pKa = 5.64CRR13 pKa = 11.84EE14 pKa = 4.41WFHH17 pKa = 6.84PARR20 pKa = 11.84EE21 pKa = 4.26GQVVCSFEE29 pKa = 4.16CASEE33 pKa = 3.93IGKK36 pKa = 8.97KK37 pKa = 7.03QTAKK41 pKa = 10.56ARR43 pKa = 11.84EE44 pKa = 3.93AAKK47 pKa = 10.47ARR49 pKa = 11.84AVKK52 pKa = 10.09RR53 pKa = 11.84QRR55 pKa = 11.84EE56 pKa = 4.09FEE58 pKa = 3.97KK59 pKa = 10.49EE60 pKa = 3.49GRR62 pKa = 11.84QRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84AKK68 pKa = 10.5RR69 pKa = 11.84EE70 pKa = 3.81SFKK73 pKa = 10.93TKK75 pKa = 10.43AQWDD79 pKa = 4.06KK80 pKa = 10.73EE81 pKa = 4.19AQSAFNRR88 pKa = 11.84YY89 pKa = 8.22IRR91 pKa = 11.84IRR93 pKa = 11.84DD94 pKa = 3.38EE95 pKa = 4.24GKK97 pKa = 9.48PCVSCGNPLIGKK109 pKa = 9.17SNYY112 pKa = 8.55LTGSAIDD119 pKa = 3.57ASHH122 pKa = 5.58YY123 pKa = 9.46RR124 pKa = 11.84SRR126 pKa = 11.84GAASHH131 pKa = 6.67LKK133 pKa = 9.12FNVFNVHH140 pKa = 6.17SACTRR145 pKa = 11.84CNRR148 pKa = 11.84QLSGNAVEE156 pKa = 4.7YY157 pKa = 10.2RR158 pKa = 11.84IHH160 pKa = 7.37LIEE163 pKa = 5.58RR164 pKa = 11.84IGLDD168 pKa = 2.75RR169 pKa = 11.84VEE171 pKa = 5.12RR172 pKa = 11.84LEE174 pKa = 4.33ADD176 pKa = 3.23NEE178 pKa = 3.95LRR180 pKa = 11.84RR181 pKa = 11.84FDD183 pKa = 3.47IPYY186 pKa = 9.14LQRR189 pKa = 11.84IKK191 pKa = 11.0SIFTRR196 pKa = 11.84RR197 pKa = 11.84ARR199 pKa = 11.84ALEE202 pKa = 3.86KK203 pKa = 10.4RR204 pKa = 11.84RR205 pKa = 11.84ARR207 pKa = 11.84HH208 pKa = 5.15QEE210 pKa = 3.52AAA212 pKa = 3.66
MM1 pKa = 7.89RR2 pKa = 11.84KK3 pKa = 8.26PARR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.32CAHH11 pKa = 5.64CRR13 pKa = 11.84EE14 pKa = 4.41WFHH17 pKa = 6.84PARR20 pKa = 11.84EE21 pKa = 4.26GQVVCSFEE29 pKa = 4.16CASEE33 pKa = 3.93IGKK36 pKa = 8.97KK37 pKa = 7.03QTAKK41 pKa = 10.56ARR43 pKa = 11.84EE44 pKa = 3.93AAKK47 pKa = 10.47ARR49 pKa = 11.84AVKK52 pKa = 10.09RR53 pKa = 11.84QRR55 pKa = 11.84EE56 pKa = 4.09FEE58 pKa = 3.97KK59 pKa = 10.49EE60 pKa = 3.49GRR62 pKa = 11.84QRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84AKK68 pKa = 10.5RR69 pKa = 11.84EE70 pKa = 3.81SFKK73 pKa = 10.93TKK75 pKa = 10.43AQWDD79 pKa = 4.06KK80 pKa = 10.73EE81 pKa = 4.19AQSAFNRR88 pKa = 11.84YY89 pKa = 8.22IRR91 pKa = 11.84IRR93 pKa = 11.84DD94 pKa = 3.38EE95 pKa = 4.24GKK97 pKa = 9.48PCVSCGNPLIGKK109 pKa = 9.17SNYY112 pKa = 8.55LTGSAIDD119 pKa = 3.57ASHH122 pKa = 5.58YY123 pKa = 9.46RR124 pKa = 11.84SRR126 pKa = 11.84GAASHH131 pKa = 6.67LKK133 pKa = 9.12FNVFNVHH140 pKa = 6.17SACTRR145 pKa = 11.84CNRR148 pKa = 11.84QLSGNAVEE156 pKa = 4.7YY157 pKa = 10.2RR158 pKa = 11.84IHH160 pKa = 7.37LIEE163 pKa = 5.58RR164 pKa = 11.84IGLDD168 pKa = 2.75RR169 pKa = 11.84VEE171 pKa = 5.12RR172 pKa = 11.84LEE174 pKa = 4.33ADD176 pKa = 3.23NEE178 pKa = 3.95LRR180 pKa = 11.84RR181 pKa = 11.84FDD183 pKa = 3.47IPYY186 pKa = 9.14LQRR189 pKa = 11.84IKK191 pKa = 11.0SIFTRR196 pKa = 11.84RR197 pKa = 11.84ARR199 pKa = 11.84ALEE202 pKa = 3.86KK203 pKa = 10.4RR204 pKa = 11.84RR205 pKa = 11.84ARR207 pKa = 11.84HH208 pKa = 5.15QEE210 pKa = 3.52AAA212 pKa = 3.66
Molecular weight: 24.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13865 |
36 |
1022 |
203.9 |
22.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.657 ± 0.37 | 1.118 ± 0.133 |
5.691 ± 0.212 | 5.604 ± 0.276 |
3.599 ± 0.257 | 7.169 ± 0.334 |
1.608 ± 0.146 | 5.979 ± 0.259 |
5.193 ± 0.412 | 9.203 ± 0.386 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.431 ± 0.139 | 3.924 ± 0.236 |
4.183 ± 0.247 | 4.342 ± 0.442 |
6.066 ± 0.322 | 6.542 ± 0.244 |
6.268 ± 0.438 | 6.686 ± 0.286 |
1.717 ± 0.145 | 3.022 ± 0.171 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
