
Black currant nucleorhabdovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus; unclassified Nucleorhabdovirus
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
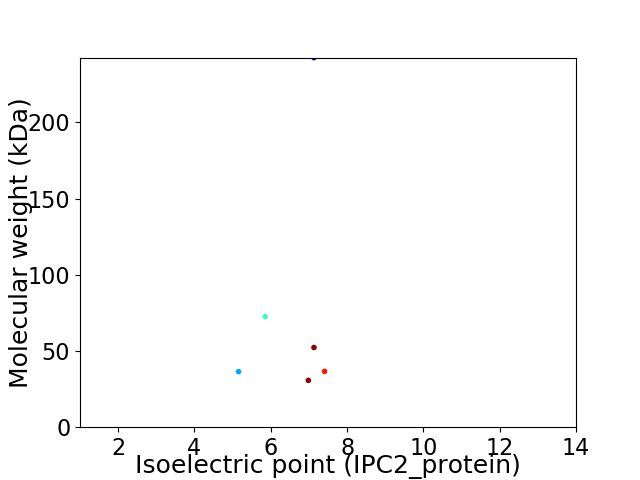
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K9YRH6|A0A2K9YRH6_9RHAB Putative cell-to-cell movement protein OS=Black currant nucleorhabdovirus 1 OX=2079521 PE=4 SV=1
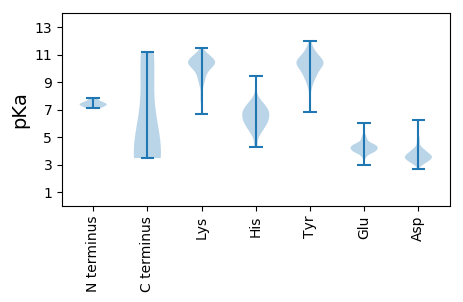
MM1 pKa = 7.48NIAGGIANVHH11 pKa = 5.92GPYY14 pKa = 10.14SGLPQSALNSEE25 pKa = 4.57VLTSRR30 pKa = 11.84LANEE34 pKa = 4.34VVKK37 pKa = 10.91SDD39 pKa = 4.06TIDD42 pKa = 3.66DD43 pKa = 3.98EE44 pKa = 4.56EE45 pKa = 4.13RR46 pKa = 11.84TIAYY50 pKa = 7.91MKK52 pKa = 10.45EE53 pKa = 3.63WMKK56 pKa = 10.87TMSDD60 pKa = 3.12KK61 pKa = 11.25GINIPTRR68 pKa = 11.84VIEE71 pKa = 4.11VLAEE75 pKa = 3.66ISIVLEE81 pKa = 3.81NSGMDD86 pKa = 3.74DD87 pKa = 3.29LHH89 pKa = 7.59GKK91 pKa = 10.09FGFAMVEE98 pKa = 4.41FGTHH102 pKa = 5.6LMKK105 pKa = 11.0NQIAPNTLLEE115 pKa = 4.34KK116 pKa = 9.01MTGLVDD122 pKa = 3.6KK123 pKa = 10.64LYY125 pKa = 11.09EE126 pKa = 4.48GVNEE130 pKa = 4.12MKK132 pKa = 9.55KK133 pKa = 9.03TNDD136 pKa = 4.97KK137 pKa = 10.76INQDD141 pKa = 3.94LPRR144 pKa = 11.84TIAKK148 pKa = 10.04AKK150 pKa = 9.44RR151 pKa = 11.84QQSGKK156 pKa = 10.16AKK158 pKa = 10.4ASEE161 pKa = 4.07TSPEE165 pKa = 3.92KK166 pKa = 11.03VNNNNLSPPSTAQSNTDD183 pKa = 3.22TGVKK187 pKa = 8.81EE188 pKa = 4.22QEE190 pKa = 4.13EE191 pKa = 4.41KK192 pKa = 10.54MEE194 pKa = 4.26TDD196 pKa = 4.26APALNPKK203 pKa = 9.09SEE205 pKa = 4.17KK206 pKa = 10.52AKK208 pKa = 9.9QLRR211 pKa = 11.84DD212 pKa = 3.39TYY214 pKa = 9.78KK215 pKa = 10.72AHH217 pKa = 6.64YY218 pKa = 7.12STPGFDD224 pKa = 3.76AYY226 pKa = 9.84TPEE229 pKa = 4.33KK230 pKa = 10.41VRR232 pKa = 11.84EE233 pKa = 4.32LITYY237 pKa = 6.85YY238 pKa = 10.65ARR240 pKa = 11.84HH241 pKa = 6.32ILDD244 pKa = 3.55IRR246 pKa = 11.84PDD248 pKa = 2.94IWQDD252 pKa = 3.29DD253 pKa = 3.64ASTFNMVSDD262 pKa = 5.54LIDD265 pKa = 3.57KK266 pKa = 10.46KK267 pKa = 11.01RR268 pKa = 11.84LFIVLSKK275 pKa = 10.88AKK277 pKa = 10.72DD278 pKa = 3.2MGLSYY283 pKa = 10.43EE284 pKa = 4.24DD285 pKa = 3.98HH286 pKa = 5.68ATARR290 pKa = 11.84EE291 pKa = 3.93EE292 pKa = 4.89FIDD295 pKa = 5.55AINTCSPAYY304 pKa = 10.1GSYY307 pKa = 8.98TVQGMIHH314 pKa = 6.98GDD316 pKa = 3.83DD317 pKa = 3.34VWIEE321 pKa = 4.06LIKK324 pKa = 10.79LDD326 pKa = 4.17
MM1 pKa = 7.48NIAGGIANVHH11 pKa = 5.92GPYY14 pKa = 10.14SGLPQSALNSEE25 pKa = 4.57VLTSRR30 pKa = 11.84LANEE34 pKa = 4.34VVKK37 pKa = 10.91SDD39 pKa = 4.06TIDD42 pKa = 3.66DD43 pKa = 3.98EE44 pKa = 4.56EE45 pKa = 4.13RR46 pKa = 11.84TIAYY50 pKa = 7.91MKK52 pKa = 10.45EE53 pKa = 3.63WMKK56 pKa = 10.87TMSDD60 pKa = 3.12KK61 pKa = 11.25GINIPTRR68 pKa = 11.84VIEE71 pKa = 4.11VLAEE75 pKa = 3.66ISIVLEE81 pKa = 3.81NSGMDD86 pKa = 3.74DD87 pKa = 3.29LHH89 pKa = 7.59GKK91 pKa = 10.09FGFAMVEE98 pKa = 4.41FGTHH102 pKa = 5.6LMKK105 pKa = 11.0NQIAPNTLLEE115 pKa = 4.34KK116 pKa = 9.01MTGLVDD122 pKa = 3.6KK123 pKa = 10.64LYY125 pKa = 11.09EE126 pKa = 4.48GVNEE130 pKa = 4.12MKK132 pKa = 9.55KK133 pKa = 9.03TNDD136 pKa = 4.97KK137 pKa = 10.76INQDD141 pKa = 3.94LPRR144 pKa = 11.84TIAKK148 pKa = 10.04AKK150 pKa = 9.44RR151 pKa = 11.84QQSGKK156 pKa = 10.16AKK158 pKa = 10.4ASEE161 pKa = 4.07TSPEE165 pKa = 3.92KK166 pKa = 11.03VNNNNLSPPSTAQSNTDD183 pKa = 3.22TGVKK187 pKa = 8.81EE188 pKa = 4.22QEE190 pKa = 4.13EE191 pKa = 4.41KK192 pKa = 10.54MEE194 pKa = 4.26TDD196 pKa = 4.26APALNPKK203 pKa = 9.09SEE205 pKa = 4.17KK206 pKa = 10.52AKK208 pKa = 9.9QLRR211 pKa = 11.84DD212 pKa = 3.39TYY214 pKa = 9.78KK215 pKa = 10.72AHH217 pKa = 6.64YY218 pKa = 7.12STPGFDD224 pKa = 3.76AYY226 pKa = 9.84TPEE229 pKa = 4.33KK230 pKa = 10.41VRR232 pKa = 11.84EE233 pKa = 4.32LITYY237 pKa = 6.85YY238 pKa = 10.65ARR240 pKa = 11.84HH241 pKa = 6.32ILDD244 pKa = 3.55IRR246 pKa = 11.84PDD248 pKa = 2.94IWQDD252 pKa = 3.29DD253 pKa = 3.64ASTFNMVSDD262 pKa = 5.54LIDD265 pKa = 3.57KK266 pKa = 10.46KK267 pKa = 11.01RR268 pKa = 11.84LFIVLSKK275 pKa = 10.88AKK277 pKa = 10.72DD278 pKa = 3.2MGLSYY283 pKa = 10.43EE284 pKa = 4.24DD285 pKa = 3.98HH286 pKa = 5.68ATARR290 pKa = 11.84EE291 pKa = 3.93EE292 pKa = 4.89FIDD295 pKa = 5.55AINTCSPAYY304 pKa = 10.1GSYY307 pKa = 8.98TVQGMIHH314 pKa = 6.98GDD316 pKa = 3.83DD317 pKa = 3.34VWIEE321 pKa = 4.06LIKK324 pKa = 10.79LDD326 pKa = 4.17
Molecular weight: 36.44 kDa
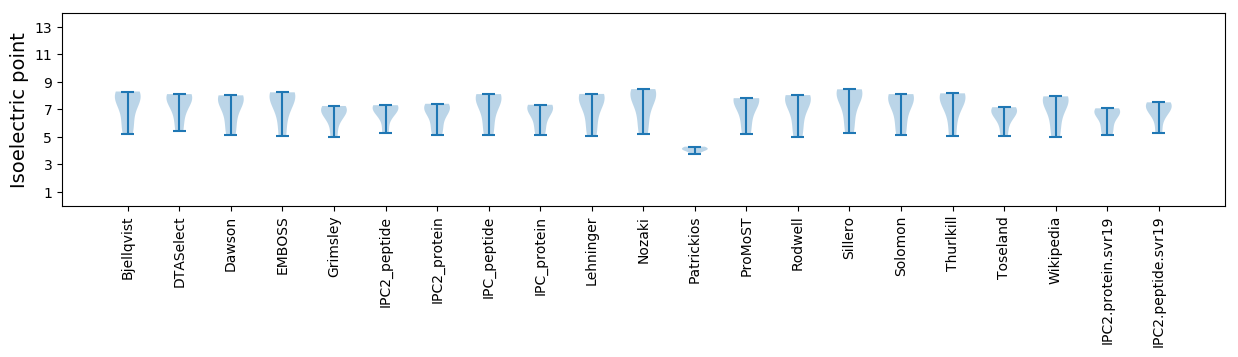
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K9YRI2|A0A2K9YRI2_9RHAB Matrix protein OS=Black currant nucleorhabdovirus 1 OX=2079521 PE=4 SV=1
MM1 pKa = 7.38TSSKK5 pKa = 9.51TRR7 pKa = 11.84FSEE10 pKa = 4.17EE11 pKa = 4.19FPSKK15 pKa = 7.42PTKK18 pKa = 9.16YY19 pKa = 9.46VSYY22 pKa = 9.75VAVPIRR28 pKa = 11.84GDD30 pKa = 3.18HH31 pKa = 5.56TEE33 pKa = 4.15IPVSADD39 pKa = 2.79GFLLSTKK46 pKa = 10.35AILYY50 pKa = 9.56IKK52 pKa = 10.37NLLSLSSLEE61 pKa = 3.95VDD63 pKa = 3.86VKK65 pKa = 10.75QVSLAWCPLVEE76 pKa = 4.52PNHH79 pKa = 6.22LSSVTIKK86 pKa = 10.52VNYY89 pKa = 9.89DD90 pKa = 3.01IAHH93 pKa = 6.44GSMLQEE99 pKa = 4.55KK100 pKa = 9.97DD101 pKa = 3.32LSLVRR106 pKa = 11.84LTGKK110 pKa = 10.04ISEE113 pKa = 3.99QHH115 pKa = 6.65GIVLYY120 pKa = 6.98PTRR123 pKa = 11.84PLLYY127 pKa = 9.72PKK129 pKa = 10.24GGKK132 pKa = 8.94LRR134 pKa = 11.84LPWDD138 pKa = 3.62VQVEE142 pKa = 3.93IDD144 pKa = 4.01GVSEE148 pKa = 4.08GSNVDD153 pKa = 3.27QTLGIMKK160 pKa = 8.33VWCQIKK166 pKa = 8.85TKK168 pKa = 10.2PFLSSNTYY176 pKa = 9.7PMKK179 pKa = 10.46LYY181 pKa = 10.22KK182 pKa = 10.51APEE185 pKa = 4.09IKK187 pKa = 9.36WGNVHH192 pKa = 6.62FPFYY196 pKa = 10.36VPYY199 pKa = 10.7YY200 pKa = 9.49IINNVRR206 pKa = 11.84GKK208 pKa = 9.22TSMEE212 pKa = 3.87WEE214 pKa = 3.54NSYY217 pKa = 10.83QYY219 pKa = 11.59EE220 pKa = 4.46LFTKK224 pKa = 10.39VVLDD228 pKa = 3.74HH229 pKa = 6.82VEE231 pKa = 4.02KK232 pKa = 11.02KK233 pKa = 10.78LINQEE238 pKa = 3.56NTLALMQTMSFIDD251 pKa = 3.52CTSINEE257 pKa = 4.1LTKK260 pKa = 11.17NCVLNKK266 pKa = 9.94NGEE269 pKa = 4.17YY270 pKa = 10.01CSCGNRR276 pKa = 11.84VKK278 pKa = 10.72DD279 pKa = 3.7YY280 pKa = 11.84VEE282 pKa = 4.21ICMKK286 pKa = 10.75DD287 pKa = 3.57RR288 pKa = 11.84DD289 pKa = 4.27DD290 pKa = 4.04MYY292 pKa = 10.76TNHH295 pKa = 6.26SSIYY299 pKa = 10.23FEE301 pKa = 4.18TTNGIKK307 pKa = 10.13KK308 pKa = 10.45GLLKK312 pKa = 10.61SSNGARR318 pKa = 11.84VPEE321 pKa = 4.28FF322 pKa = 3.47
MM1 pKa = 7.38TSSKK5 pKa = 9.51TRR7 pKa = 11.84FSEE10 pKa = 4.17EE11 pKa = 4.19FPSKK15 pKa = 7.42PTKK18 pKa = 9.16YY19 pKa = 9.46VSYY22 pKa = 9.75VAVPIRR28 pKa = 11.84GDD30 pKa = 3.18HH31 pKa = 5.56TEE33 pKa = 4.15IPVSADD39 pKa = 2.79GFLLSTKK46 pKa = 10.35AILYY50 pKa = 9.56IKK52 pKa = 10.37NLLSLSSLEE61 pKa = 3.95VDD63 pKa = 3.86VKK65 pKa = 10.75QVSLAWCPLVEE76 pKa = 4.52PNHH79 pKa = 6.22LSSVTIKK86 pKa = 10.52VNYY89 pKa = 9.89DD90 pKa = 3.01IAHH93 pKa = 6.44GSMLQEE99 pKa = 4.55KK100 pKa = 9.97DD101 pKa = 3.32LSLVRR106 pKa = 11.84LTGKK110 pKa = 10.04ISEE113 pKa = 3.99QHH115 pKa = 6.65GIVLYY120 pKa = 6.98PTRR123 pKa = 11.84PLLYY127 pKa = 9.72PKK129 pKa = 10.24GGKK132 pKa = 8.94LRR134 pKa = 11.84LPWDD138 pKa = 3.62VQVEE142 pKa = 3.93IDD144 pKa = 4.01GVSEE148 pKa = 4.08GSNVDD153 pKa = 3.27QTLGIMKK160 pKa = 8.33VWCQIKK166 pKa = 8.85TKK168 pKa = 10.2PFLSSNTYY176 pKa = 9.7PMKK179 pKa = 10.46LYY181 pKa = 10.22KK182 pKa = 10.51APEE185 pKa = 4.09IKK187 pKa = 9.36WGNVHH192 pKa = 6.62FPFYY196 pKa = 10.36VPYY199 pKa = 10.7YY200 pKa = 9.49IINNVRR206 pKa = 11.84GKK208 pKa = 9.22TSMEE212 pKa = 3.87WEE214 pKa = 3.54NSYY217 pKa = 10.83QYY219 pKa = 11.59EE220 pKa = 4.46LFTKK224 pKa = 10.39VVLDD228 pKa = 3.74HH229 pKa = 6.82VEE231 pKa = 4.02KK232 pKa = 11.02KK233 pKa = 10.78LINQEE238 pKa = 3.56NTLALMQTMSFIDD251 pKa = 3.52CTSINEE257 pKa = 4.1LTKK260 pKa = 11.17NCVLNKK266 pKa = 9.94NGEE269 pKa = 4.17YY270 pKa = 10.01CSCGNRR276 pKa = 11.84VKK278 pKa = 10.72DD279 pKa = 3.7YY280 pKa = 11.84VEE282 pKa = 4.21ICMKK286 pKa = 10.75DD287 pKa = 3.57RR288 pKa = 11.84DD289 pKa = 4.27DD290 pKa = 4.04MYY292 pKa = 10.76TNHH295 pKa = 6.26SSIYY299 pKa = 10.23FEE301 pKa = 4.18TTNGIKK307 pKa = 10.13KK308 pKa = 10.45GLLKK312 pKa = 10.61SSNGARR318 pKa = 11.84VPEE321 pKa = 4.28FF322 pKa = 3.47
Molecular weight: 36.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4139 |
276 |
2106 |
689.8 |
78.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.856 ± 1.132 | 1.957 ± 0.308 |
6.185 ± 0.441 | 5.654 ± 0.357 |
3.6 ± 0.224 | 5.025 ± 0.244 |
2.561 ± 0.414 | 7.466 ± 0.504 |
7.345 ± 0.462 | 9.133 ± 0.802 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.213 ± 0.109 | 4.349 ± 0.306 |
4.373 ± 0.748 | 3.141 ± 0.16 |
4.421 ± 0.607 | 9.181 ± 0.658 |
6.282 ± 0.527 | 5.146 ± 0.368 |
1.208 ± 0.2 | 4.905 ± 0.202 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
