
Oat chlorotic stunt virus (isolate United Kingdom) (OCSV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Avenavirus; Oat chlorotic stunt virus
Average proteome isoelectric point is 8.37
Get precalculated fractions of proteins
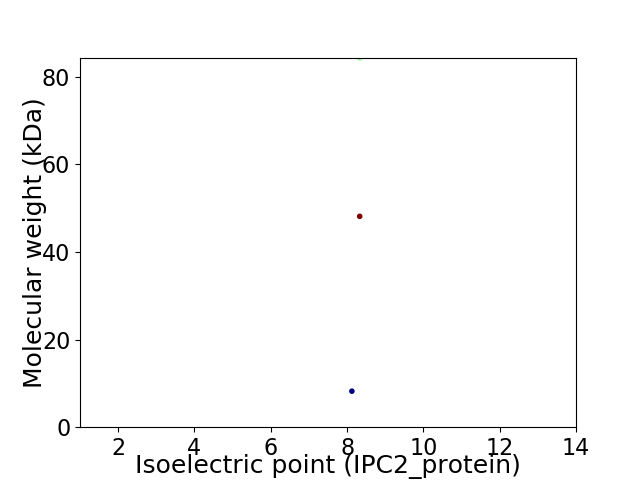
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q83929|P8_OCSVU Uncharacterized protein p8 OS=Oat chlorotic stunt virus (isolate United Kingdom) OX=652110 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.68RR2 pKa = 11.84WLSRR6 pKa = 11.84TVKK9 pKa = 10.16CWVPLILAPLHH20 pKa = 6.38RR21 pKa = 11.84MSPLSVALATEE32 pKa = 5.51LILGCQLSSLGCLQLPLITRR52 pKa = 11.84STNFVGSASLLSRR65 pKa = 11.84WFLPIIAEE73 pKa = 4.19EE74 pKa = 4.04
MM1 pKa = 7.68RR2 pKa = 11.84WLSRR6 pKa = 11.84TVKK9 pKa = 10.16CWVPLILAPLHH20 pKa = 6.38RR21 pKa = 11.84MSPLSVALATEE32 pKa = 5.51LILGCQLSSLGCLQLPLITRR52 pKa = 11.84STNFVGSASLLSRR65 pKa = 11.84WFLPIIAEE73 pKa = 4.19EE74 pKa = 4.04
Molecular weight: 8.22 kDa
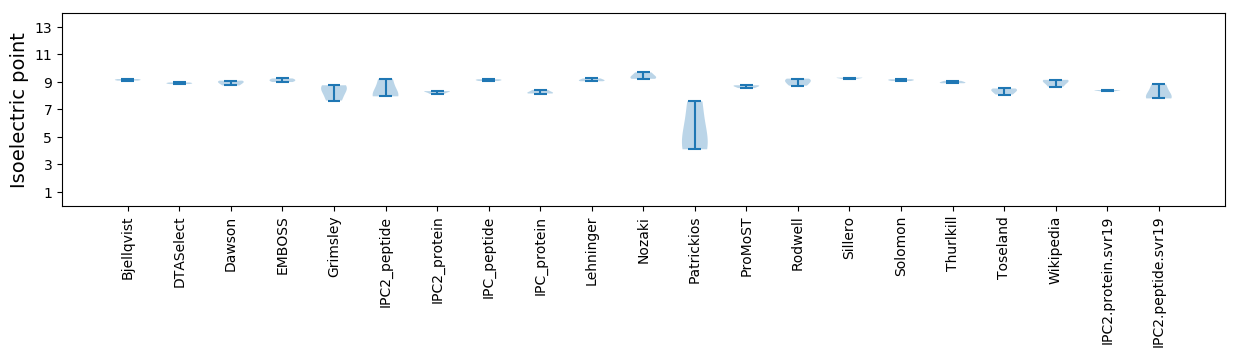
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q83928|CAPSD_OCSVU Capsid protein OS=Oat chlorotic stunt virus (isolate United Kingdom) OX=652110 GN=ORF2 PE=3 SV=1
MM1 pKa = 7.49SWPSLSGVPNGSSSVSTWLSVTHH24 pKa = 6.37PRR26 pKa = 11.84RR27 pKa = 11.84SLPKK31 pKa = 9.65EE32 pKa = 3.69LLTAFDD38 pKa = 3.97TCNVAPEE45 pKa = 4.16ALLVLRR51 pKa = 11.84STSLMILEE59 pKa = 4.38EE60 pKa = 4.24TCVVVGAAEE69 pKa = 4.13MPTAEE74 pKa = 4.62DD75 pKa = 3.31NSGRR79 pKa = 11.84EE80 pKa = 4.1LFIGSNGDD88 pKa = 3.08PMEE91 pKa = 4.67RR92 pKa = 11.84KK93 pKa = 7.71TRR95 pKa = 11.84TAHH98 pKa = 5.87HH99 pKa = 7.14AIKK102 pKa = 9.36KK103 pKa = 5.84TVRR106 pKa = 11.84IKK108 pKa = 10.48KK109 pKa = 7.85GHH111 pKa = 5.79RR112 pKa = 11.84TTFAMTVANGAYY124 pKa = 9.69VKK126 pKa = 10.69FGARR130 pKa = 11.84PLTEE134 pKa = 4.45ANVLVVRR141 pKa = 11.84KK142 pKa = 9.44WIVKK146 pKa = 9.94LIADD150 pKa = 4.41EE151 pKa = 4.33YY152 pKa = 11.2KK153 pKa = 10.61DD154 pKa = 5.61LRR156 pKa = 11.84VCDD159 pKa = 3.51QALVIDD165 pKa = 4.49RR166 pKa = 11.84ATFLSFIPTMAWNNYY181 pKa = 8.53KK182 pKa = 10.66FIFHH186 pKa = 6.98GKK188 pKa = 8.61NAVTDD193 pKa = 4.26RR194 pKa = 11.84VAGEE198 pKa = 4.06NLFSRR203 pKa = 11.84IAQWANPGKK212 pKa = 7.85XGCPVVVTGQGCVISRR228 pKa = 11.84APDD231 pKa = 3.54CAQLRR236 pKa = 11.84VKK238 pKa = 10.59RR239 pKa = 11.84LLGVTKK245 pKa = 10.48NRR247 pKa = 11.84TCMRR251 pKa = 11.84VSGVSPNIQIIPFNNDD267 pKa = 1.5ITTLEE272 pKa = 3.95RR273 pKa = 11.84AIKK276 pKa = 10.1EE277 pKa = 3.67RR278 pKa = 11.84VFFVKK283 pKa = 10.52NLDD286 pKa = 3.47KK287 pKa = 11.15GSPTKK292 pKa = 10.3FVSPPRR298 pKa = 11.84PAPGVFAQRR307 pKa = 11.84LSNTLGLLVPFLPSTAPMSHH327 pKa = 5.09QQFVDD332 pKa = 3.51STPSRR337 pKa = 11.84KK338 pKa = 9.59RR339 pKa = 11.84KK340 pKa = 9.58VYY342 pKa = 9.88QQALEE347 pKa = 5.08DD348 pKa = 4.04ISCHH352 pKa = 5.09GLNLEE357 pKa = 4.14TDD359 pKa = 3.77SKK361 pKa = 11.55VKK363 pKa = 10.8VFVKK367 pKa = 10.33YY368 pKa = 10.48EE369 pKa = 4.19KK370 pKa = 9.81TDD372 pKa = 3.44HH373 pKa = 6.22TSKK376 pKa = 10.94ADD378 pKa = 3.39PVPRR382 pKa = 11.84VISPRR387 pKa = 11.84DD388 pKa = 3.33PKK390 pKa = 11.46YY391 pKa = 10.65NLALGRR397 pKa = 11.84YY398 pKa = 6.96LRR400 pKa = 11.84PMEE403 pKa = 3.9EE404 pKa = 4.57RR405 pKa = 11.84IFKK408 pKa = 10.79ALGKK412 pKa = 10.23LFGHH416 pKa = 5.7RR417 pKa = 11.84TVMKK421 pKa = 10.77GMDD424 pKa = 3.57TDD426 pKa = 3.39VTARR430 pKa = 11.84VIQEE434 pKa = 3.64KK435 pKa = 10.16WNMFNKK441 pKa = 9.85PVAIGLDD448 pKa = 3.26ASRR451 pKa = 11.84FDD453 pKa = 3.31QHH455 pKa = 7.58VSLEE459 pKa = 4.06ALEE462 pKa = 4.92FEE464 pKa = 4.21HH465 pKa = 7.49SVYY468 pKa = 10.45LKK470 pKa = 10.21CVRR473 pKa = 11.84RR474 pKa = 11.84MVDD477 pKa = 2.93KK478 pKa = 11.0RR479 pKa = 11.84KK480 pKa = 10.14LGNILRR486 pKa = 11.84HH487 pKa = 5.24QLLNKK492 pKa = 10.3CYY494 pKa = 10.78GNTPDD499 pKa = 4.16GAVSYY504 pKa = 9.09TIEE507 pKa = 4.19GTRR510 pKa = 11.84MSGDD514 pKa = 3.41MNTSLGNCVLMCMMIHH530 pKa = 7.54AYY532 pKa = 9.94GLHH535 pKa = 6.25KK536 pKa = 10.63SVNIQLANNGDD547 pKa = 3.8DD548 pKa = 3.28CVVFLEE554 pKa = 4.22QSDD557 pKa = 4.08LATFSEE563 pKa = 5.53GLFEE567 pKa = 4.6WFLEE571 pKa = 4.15MGFNMAIEE579 pKa = 4.37EE580 pKa = 4.0PSYY583 pKa = 10.47EE584 pKa = 3.98LEE586 pKa = 4.77HH587 pKa = 8.84IEE589 pKa = 4.13FCQCRR594 pKa = 11.84PVFDD598 pKa = 3.85GVKK601 pKa = 9.18YY602 pKa = 8.6TMCRR606 pKa = 11.84NPRR609 pKa = 11.84TAIAKK614 pKa = 9.78DD615 pKa = 3.53SVYY618 pKa = 10.64LKK620 pKa = 10.75HH621 pKa = 6.66VDD623 pKa = 3.41QFVTYY628 pKa = 10.05SSWLNAVGTGGLALAGGLPIFDD650 pKa = 5.14AFYY653 pKa = 10.05TCYY656 pKa = 10.39KK657 pKa = 10.17RR658 pKa = 11.84NSNSHH663 pKa = 5.22WFSGRR668 pKa = 11.84KK669 pKa = 9.2GRR671 pKa = 11.84LKK673 pKa = 9.97TLSSVDD679 pKa = 4.78DD680 pKa = 3.78SLPWFMRR687 pKa = 11.84EE688 pKa = 4.12LGLKK692 pKa = 9.72GKK694 pKa = 10.25RR695 pKa = 11.84SSAEE699 pKa = 3.61PLPASRR705 pKa = 11.84ASFYY709 pKa = 10.74LAWGVTPCEE718 pKa = 3.68QLEE721 pKa = 4.18LEE723 pKa = 4.61KK724 pKa = 10.86YY725 pKa = 9.31YY726 pKa = 11.11KK727 pKa = 10.29SFKK730 pKa = 10.61LDD732 pKa = 3.03TSTLLEE738 pKa = 3.81EE739 pKa = 5.48HH740 pKa = 6.77LWQPRR745 pKa = 11.84GVFPDD750 pKa = 3.38EE751 pKa = 4.4DD752 pKa = 3.52
MM1 pKa = 7.49SWPSLSGVPNGSSSVSTWLSVTHH24 pKa = 6.37PRR26 pKa = 11.84RR27 pKa = 11.84SLPKK31 pKa = 9.65EE32 pKa = 3.69LLTAFDD38 pKa = 3.97TCNVAPEE45 pKa = 4.16ALLVLRR51 pKa = 11.84STSLMILEE59 pKa = 4.38EE60 pKa = 4.24TCVVVGAAEE69 pKa = 4.13MPTAEE74 pKa = 4.62DD75 pKa = 3.31NSGRR79 pKa = 11.84EE80 pKa = 4.1LFIGSNGDD88 pKa = 3.08PMEE91 pKa = 4.67RR92 pKa = 11.84KK93 pKa = 7.71TRR95 pKa = 11.84TAHH98 pKa = 5.87HH99 pKa = 7.14AIKK102 pKa = 9.36KK103 pKa = 5.84TVRR106 pKa = 11.84IKK108 pKa = 10.48KK109 pKa = 7.85GHH111 pKa = 5.79RR112 pKa = 11.84TTFAMTVANGAYY124 pKa = 9.69VKK126 pKa = 10.69FGARR130 pKa = 11.84PLTEE134 pKa = 4.45ANVLVVRR141 pKa = 11.84KK142 pKa = 9.44WIVKK146 pKa = 9.94LIADD150 pKa = 4.41EE151 pKa = 4.33YY152 pKa = 11.2KK153 pKa = 10.61DD154 pKa = 5.61LRR156 pKa = 11.84VCDD159 pKa = 3.51QALVIDD165 pKa = 4.49RR166 pKa = 11.84ATFLSFIPTMAWNNYY181 pKa = 8.53KK182 pKa = 10.66FIFHH186 pKa = 6.98GKK188 pKa = 8.61NAVTDD193 pKa = 4.26RR194 pKa = 11.84VAGEE198 pKa = 4.06NLFSRR203 pKa = 11.84IAQWANPGKK212 pKa = 7.85XGCPVVVTGQGCVISRR228 pKa = 11.84APDD231 pKa = 3.54CAQLRR236 pKa = 11.84VKK238 pKa = 10.59RR239 pKa = 11.84LLGVTKK245 pKa = 10.48NRR247 pKa = 11.84TCMRR251 pKa = 11.84VSGVSPNIQIIPFNNDD267 pKa = 1.5ITTLEE272 pKa = 3.95RR273 pKa = 11.84AIKK276 pKa = 10.1EE277 pKa = 3.67RR278 pKa = 11.84VFFVKK283 pKa = 10.52NLDD286 pKa = 3.47KK287 pKa = 11.15GSPTKK292 pKa = 10.3FVSPPRR298 pKa = 11.84PAPGVFAQRR307 pKa = 11.84LSNTLGLLVPFLPSTAPMSHH327 pKa = 5.09QQFVDD332 pKa = 3.51STPSRR337 pKa = 11.84KK338 pKa = 9.59RR339 pKa = 11.84KK340 pKa = 9.58VYY342 pKa = 9.88QQALEE347 pKa = 5.08DD348 pKa = 4.04ISCHH352 pKa = 5.09GLNLEE357 pKa = 4.14TDD359 pKa = 3.77SKK361 pKa = 11.55VKK363 pKa = 10.8VFVKK367 pKa = 10.33YY368 pKa = 10.48EE369 pKa = 4.19KK370 pKa = 9.81TDD372 pKa = 3.44HH373 pKa = 6.22TSKK376 pKa = 10.94ADD378 pKa = 3.39PVPRR382 pKa = 11.84VISPRR387 pKa = 11.84DD388 pKa = 3.33PKK390 pKa = 11.46YY391 pKa = 10.65NLALGRR397 pKa = 11.84YY398 pKa = 6.96LRR400 pKa = 11.84PMEE403 pKa = 3.9EE404 pKa = 4.57RR405 pKa = 11.84IFKK408 pKa = 10.79ALGKK412 pKa = 10.23LFGHH416 pKa = 5.7RR417 pKa = 11.84TVMKK421 pKa = 10.77GMDD424 pKa = 3.57TDD426 pKa = 3.39VTARR430 pKa = 11.84VIQEE434 pKa = 3.64KK435 pKa = 10.16WNMFNKK441 pKa = 9.85PVAIGLDD448 pKa = 3.26ASRR451 pKa = 11.84FDD453 pKa = 3.31QHH455 pKa = 7.58VSLEE459 pKa = 4.06ALEE462 pKa = 4.92FEE464 pKa = 4.21HH465 pKa = 7.49SVYY468 pKa = 10.45LKK470 pKa = 10.21CVRR473 pKa = 11.84RR474 pKa = 11.84MVDD477 pKa = 2.93KK478 pKa = 11.0RR479 pKa = 11.84KK480 pKa = 10.14LGNILRR486 pKa = 11.84HH487 pKa = 5.24QLLNKK492 pKa = 10.3CYY494 pKa = 10.78GNTPDD499 pKa = 4.16GAVSYY504 pKa = 9.09TIEE507 pKa = 4.19GTRR510 pKa = 11.84MSGDD514 pKa = 3.41MNTSLGNCVLMCMMIHH530 pKa = 7.54AYY532 pKa = 9.94GLHH535 pKa = 6.25KK536 pKa = 10.63SVNIQLANNGDD547 pKa = 3.8DD548 pKa = 3.28CVVFLEE554 pKa = 4.22QSDD557 pKa = 4.08LATFSEE563 pKa = 5.53GLFEE567 pKa = 4.6WFLEE571 pKa = 4.15MGFNMAIEE579 pKa = 4.37EE580 pKa = 4.0PSYY583 pKa = 10.47EE584 pKa = 3.98LEE586 pKa = 4.77HH587 pKa = 8.84IEE589 pKa = 4.13FCQCRR594 pKa = 11.84PVFDD598 pKa = 3.85GVKK601 pKa = 9.18YY602 pKa = 8.6TMCRR606 pKa = 11.84NPRR609 pKa = 11.84TAIAKK614 pKa = 9.78DD615 pKa = 3.53SVYY618 pKa = 10.64LKK620 pKa = 10.75HH621 pKa = 6.66VDD623 pKa = 3.41QFVTYY628 pKa = 10.05SSWLNAVGTGGLALAGGLPIFDD650 pKa = 5.14AFYY653 pKa = 10.05TCYY656 pKa = 10.39KK657 pKa = 10.17RR658 pKa = 11.84NSNSHH663 pKa = 5.22WFSGRR668 pKa = 11.84KK669 pKa = 9.2GRR671 pKa = 11.84LKK673 pKa = 9.97TLSSVDD679 pKa = 4.78DD680 pKa = 3.78SLPWFMRR687 pKa = 11.84EE688 pKa = 4.12LGLKK692 pKa = 9.72GKK694 pKa = 10.25RR695 pKa = 11.84SSAEE699 pKa = 3.61PLPASRR705 pKa = 11.84ASFYY709 pKa = 10.74LAWGVTPCEE718 pKa = 3.68QLEE721 pKa = 4.18LEE723 pKa = 4.61KK724 pKa = 10.86YY725 pKa = 9.31YY726 pKa = 11.11KK727 pKa = 10.29SFKK730 pKa = 10.61LDD732 pKa = 3.03TSTLLEE738 pKa = 3.81EE739 pKa = 5.48HH740 pKa = 6.77LWQPRR745 pKa = 11.84GVFPDD750 pKa = 3.38EE751 pKa = 4.4DD752 pKa = 3.52
Molecular weight: 84.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1280 |
74 |
752 |
426.7 |
46.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.187 ± 0.489 | 2.344 ± 0.246 |
3.828 ± 0.846 | 4.375 ± 0.811 |
4.297 ± 0.523 | 7.109 ± 0.848 |
2.109 ± 0.239 | 4.297 ± 0.428 |
5.391 ± 0.879 | 9.844 ± 1.519 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.656 ± 0.237 | 4.688 ± 0.509 |
5.391 ± 0.176 | 2.344 ± 0.179 |
5.391 ± 0.934 | 8.281 ± 0.95 |
7.734 ± 1.529 | 8.438 ± 0.56 |
1.406 ± 0.433 | 2.813 ± 0.435 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
