
Porcine picobirnavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Picobirnaviridae; Picobirnavirus; unclassified Picobirnavirus
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
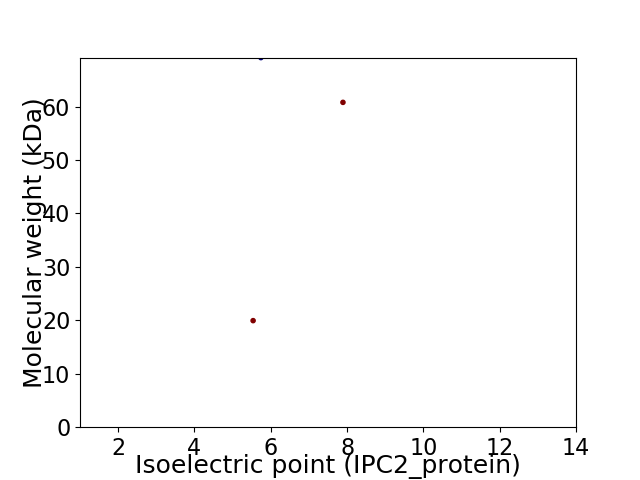
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W6B0R5|W6B0R5_9VIRU Uncharacterized protein OS=Porcine picobirnavirus OX=442302 PE=4 SV=1
MM1 pKa = 7.01ATKK4 pKa = 10.3NKK6 pKa = 9.59KK7 pKa = 10.24FNGKK11 pKa = 8.46GQGYY15 pKa = 9.39RR16 pKa = 11.84RR17 pKa = 11.84GSKK20 pKa = 10.0SRR22 pKa = 11.84SDD24 pKa = 3.79SYY26 pKa = 12.02SKK28 pKa = 10.97DD29 pKa = 3.23KK30 pKa = 11.48SDD32 pKa = 3.68FSYY35 pKa = 11.4EE36 pKa = 4.08EE37 pKa = 4.03EE38 pKa = 4.36TKK40 pKa = 10.69SGKK43 pKa = 9.5DD44 pKa = 3.1RR45 pKa = 11.84QKK47 pKa = 11.21CNSSRR52 pKa = 11.84PNDD55 pKa = 3.89WKK57 pKa = 10.52WYY59 pKa = 8.27AQNEE63 pKa = 4.11QLLRR67 pKa = 11.84DD68 pKa = 3.95SASFSYY74 pKa = 8.41NTPLGTVVRR83 pKa = 11.84RR84 pKa = 11.84TVSGDD89 pKa = 3.34QPVDD93 pKa = 3.78FAGVPGVMAIYY104 pKa = 7.5TTPAYY109 pKa = 10.12GNSDD113 pKa = 3.65SAVSPLNVAIRR124 pKa = 11.84NIYY127 pKa = 10.34SFVRR131 pKa = 11.84HH132 pKa = 6.08ANSGHH137 pKa = 6.44ANYY140 pKa = 8.96DD141 pKa = 3.51APDD144 pKa = 3.56LGIYY148 pKa = 9.76LMAMDD153 pKa = 5.54NIYY156 pKa = 11.13SFMAYY161 pKa = 9.89LRR163 pKa = 11.84RR164 pKa = 11.84IYY166 pKa = 10.03GVAMTYY172 pKa = 10.49SYY174 pKa = 10.52TNKK177 pKa = 9.6YY178 pKa = 9.68YY179 pKa = 10.24PVAVTNAMGVDD190 pKa = 4.55FEE192 pKa = 5.29DD193 pKa = 3.33VHH195 pKa = 8.38ARR197 pKa = 11.84LADD200 pKa = 4.14FRR202 pKa = 11.84AMINSFAVKK211 pKa = 9.61VGSMCIPSSMSVFARR226 pKa = 11.84HH227 pKa = 4.03MWMYY231 pKa = 10.34EE232 pKa = 3.84GYY234 pKa = 8.33YY235 pKa = 10.23TDD237 pKa = 3.4TDD239 pKa = 3.53QNKK242 pKa = 8.91AQTYY246 pKa = 9.14LYY248 pKa = 10.22VPTGFYY254 pKa = 10.19HH255 pKa = 5.76YY256 pKa = 10.58RR257 pKa = 11.84LRR259 pKa = 11.84NKK261 pKa = 10.03LVEE264 pKa = 4.24EE265 pKa = 4.49GVYY268 pKa = 9.24DD269 pKa = 3.59TSGMLWYY276 pKa = 10.38KK277 pKa = 10.6PLTPYY282 pKa = 10.7GNPVSQEE289 pKa = 3.72VSQVNLLTTEE299 pKa = 4.07MLHH302 pKa = 6.59QYY304 pKa = 9.82GEE306 pKa = 4.28AMLEE310 pKa = 3.91PVLRR314 pKa = 11.84SEE316 pKa = 5.68DD317 pKa = 3.78MNIMSGDD324 pKa = 3.25ILKK327 pKa = 10.81AFGSNGIYY335 pKa = 8.53MIPMIPEE342 pKa = 3.99NYY344 pKa = 9.37VVVPSYY350 pKa = 11.05SRR352 pKa = 11.84EE353 pKa = 3.9VLDD356 pKa = 5.18QINNCTLIGDD366 pKa = 3.97NVHH369 pKa = 5.47YY370 pKa = 9.95TEE372 pKa = 5.26EE373 pKa = 4.54ADD375 pKa = 3.59FDD377 pKa = 3.51KK378 pKa = 11.34GYY380 pKa = 11.05NVLVQDD386 pKa = 4.55PKK388 pKa = 10.81HH389 pKa = 6.46GYY391 pKa = 10.32LIGKK395 pKa = 6.4TCTEE399 pKa = 3.64IPMGVAEE406 pKa = 5.02KK407 pKa = 9.73LTLNMDD413 pKa = 4.09RR414 pKa = 11.84LTAKK418 pKa = 9.74TFVNFDD424 pKa = 3.77HH425 pKa = 7.98DD426 pKa = 4.52DD427 pKa = 3.59VTPADD432 pKa = 3.69TMVATRR438 pKa = 11.84LTNVYY443 pKa = 9.87DD444 pKa = 4.15PKK446 pKa = 10.82DD447 pKa = 3.49VSIIARR453 pKa = 11.84GIGGGAAIQIRR464 pKa = 11.84PGDD467 pKa = 3.65KK468 pKa = 10.72SITLVNTTGTDD479 pKa = 2.8IANYY483 pKa = 10.44ARR485 pKa = 11.84IFYY488 pKa = 8.8YY489 pKa = 10.95ANGGLIVSEE498 pKa = 4.72MICALLPVTLQWDD511 pKa = 4.62FTTMTTEE518 pKa = 3.6KK519 pKa = 10.72LLRR522 pKa = 11.84EE523 pKa = 4.33YY524 pKa = 10.8VSKK527 pKa = 10.94AVTSMQLDD535 pKa = 3.81TARR538 pKa = 11.84INMVTNFRR546 pKa = 11.84RR547 pKa = 11.84HH548 pKa = 5.19PRR550 pKa = 11.84IAYY553 pKa = 9.36QYY555 pKa = 9.29SLYY558 pKa = 11.13VKK560 pKa = 9.25ATADD564 pKa = 3.67GAPKK568 pKa = 9.99EE569 pKa = 4.34VNEE572 pKa = 4.14HH573 pKa = 5.13GKK575 pKa = 10.17YY576 pKa = 9.94VADD579 pKa = 5.02ISDD582 pKa = 3.98LNQWTVIGSQEE593 pKa = 4.32LCNMAEE599 pKa = 4.06TALLSEE605 pKa = 4.95FNVTQYY611 pKa = 10.76GRR613 pKa = 11.84KK614 pKa = 9.26AA615 pKa = 3.14
MM1 pKa = 7.01ATKK4 pKa = 10.3NKK6 pKa = 9.59KK7 pKa = 10.24FNGKK11 pKa = 8.46GQGYY15 pKa = 9.39RR16 pKa = 11.84RR17 pKa = 11.84GSKK20 pKa = 10.0SRR22 pKa = 11.84SDD24 pKa = 3.79SYY26 pKa = 12.02SKK28 pKa = 10.97DD29 pKa = 3.23KK30 pKa = 11.48SDD32 pKa = 3.68FSYY35 pKa = 11.4EE36 pKa = 4.08EE37 pKa = 4.03EE38 pKa = 4.36TKK40 pKa = 10.69SGKK43 pKa = 9.5DD44 pKa = 3.1RR45 pKa = 11.84QKK47 pKa = 11.21CNSSRR52 pKa = 11.84PNDD55 pKa = 3.89WKK57 pKa = 10.52WYY59 pKa = 8.27AQNEE63 pKa = 4.11QLLRR67 pKa = 11.84DD68 pKa = 3.95SASFSYY74 pKa = 8.41NTPLGTVVRR83 pKa = 11.84RR84 pKa = 11.84TVSGDD89 pKa = 3.34QPVDD93 pKa = 3.78FAGVPGVMAIYY104 pKa = 7.5TTPAYY109 pKa = 10.12GNSDD113 pKa = 3.65SAVSPLNVAIRR124 pKa = 11.84NIYY127 pKa = 10.34SFVRR131 pKa = 11.84HH132 pKa = 6.08ANSGHH137 pKa = 6.44ANYY140 pKa = 8.96DD141 pKa = 3.51APDD144 pKa = 3.56LGIYY148 pKa = 9.76LMAMDD153 pKa = 5.54NIYY156 pKa = 11.13SFMAYY161 pKa = 9.89LRR163 pKa = 11.84RR164 pKa = 11.84IYY166 pKa = 10.03GVAMTYY172 pKa = 10.49SYY174 pKa = 10.52TNKK177 pKa = 9.6YY178 pKa = 9.68YY179 pKa = 10.24PVAVTNAMGVDD190 pKa = 4.55FEE192 pKa = 5.29DD193 pKa = 3.33VHH195 pKa = 8.38ARR197 pKa = 11.84LADD200 pKa = 4.14FRR202 pKa = 11.84AMINSFAVKK211 pKa = 9.61VGSMCIPSSMSVFARR226 pKa = 11.84HH227 pKa = 4.03MWMYY231 pKa = 10.34EE232 pKa = 3.84GYY234 pKa = 8.33YY235 pKa = 10.23TDD237 pKa = 3.4TDD239 pKa = 3.53QNKK242 pKa = 8.91AQTYY246 pKa = 9.14LYY248 pKa = 10.22VPTGFYY254 pKa = 10.19HH255 pKa = 5.76YY256 pKa = 10.58RR257 pKa = 11.84LRR259 pKa = 11.84NKK261 pKa = 10.03LVEE264 pKa = 4.24EE265 pKa = 4.49GVYY268 pKa = 9.24DD269 pKa = 3.59TSGMLWYY276 pKa = 10.38KK277 pKa = 10.6PLTPYY282 pKa = 10.7GNPVSQEE289 pKa = 3.72VSQVNLLTTEE299 pKa = 4.07MLHH302 pKa = 6.59QYY304 pKa = 9.82GEE306 pKa = 4.28AMLEE310 pKa = 3.91PVLRR314 pKa = 11.84SEE316 pKa = 5.68DD317 pKa = 3.78MNIMSGDD324 pKa = 3.25ILKK327 pKa = 10.81AFGSNGIYY335 pKa = 8.53MIPMIPEE342 pKa = 3.99NYY344 pKa = 9.37VVVPSYY350 pKa = 11.05SRR352 pKa = 11.84EE353 pKa = 3.9VLDD356 pKa = 5.18QINNCTLIGDD366 pKa = 3.97NVHH369 pKa = 5.47YY370 pKa = 9.95TEE372 pKa = 5.26EE373 pKa = 4.54ADD375 pKa = 3.59FDD377 pKa = 3.51KK378 pKa = 11.34GYY380 pKa = 11.05NVLVQDD386 pKa = 4.55PKK388 pKa = 10.81HH389 pKa = 6.46GYY391 pKa = 10.32LIGKK395 pKa = 6.4TCTEE399 pKa = 3.64IPMGVAEE406 pKa = 5.02KK407 pKa = 9.73LTLNMDD413 pKa = 4.09RR414 pKa = 11.84LTAKK418 pKa = 9.74TFVNFDD424 pKa = 3.77HH425 pKa = 7.98DD426 pKa = 4.52DD427 pKa = 3.59VTPADD432 pKa = 3.69TMVATRR438 pKa = 11.84LTNVYY443 pKa = 9.87DD444 pKa = 4.15PKK446 pKa = 10.82DD447 pKa = 3.49VSIIARR453 pKa = 11.84GIGGGAAIQIRR464 pKa = 11.84PGDD467 pKa = 3.65KK468 pKa = 10.72SITLVNTTGTDD479 pKa = 2.8IANYY483 pKa = 10.44ARR485 pKa = 11.84IFYY488 pKa = 8.8YY489 pKa = 10.95ANGGLIVSEE498 pKa = 4.72MICALLPVTLQWDD511 pKa = 4.62FTTMTTEE518 pKa = 3.6KK519 pKa = 10.72LLRR522 pKa = 11.84EE523 pKa = 4.33YY524 pKa = 10.8VSKK527 pKa = 10.94AVTSMQLDD535 pKa = 3.81TARR538 pKa = 11.84INMVTNFRR546 pKa = 11.84RR547 pKa = 11.84HH548 pKa = 5.19PRR550 pKa = 11.84IAYY553 pKa = 9.36QYY555 pKa = 9.29SLYY558 pKa = 11.13VKK560 pKa = 9.25ATADD564 pKa = 3.67GAPKK568 pKa = 9.99EE569 pKa = 4.34VNEE572 pKa = 4.14HH573 pKa = 5.13GKK575 pKa = 10.17YY576 pKa = 9.94VADD579 pKa = 5.02ISDD582 pKa = 3.98LNQWTVIGSQEE593 pKa = 4.32LCNMAEE599 pKa = 4.06TALLSEE605 pKa = 4.95FNVTQYY611 pKa = 10.76GRR613 pKa = 11.84KK614 pKa = 9.26AA615 pKa = 3.14
Molecular weight: 69.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W6B0S5|W6B0S5_9VIRU RNA-dependent RNA polymerase OS=Porcine picobirnavirus OX=442302 PE=4 SV=1
MM1 pKa = 7.87PKK3 pKa = 10.47NNANKK8 pKa = 10.2RR9 pKa = 11.84SRR11 pKa = 11.84DD12 pKa = 3.55YY13 pKa = 10.54FASSFASSANLRR25 pKa = 11.84AYY27 pKa = 10.02FGSVVKK33 pKa = 10.49GQSEE37 pKa = 5.11VYY39 pKa = 8.01DD40 pKa = 3.67TPFARR45 pKa = 11.84DD46 pKa = 3.35EE47 pKa = 4.27STKK50 pKa = 10.55TLLSKK55 pKa = 10.1WKK57 pKa = 10.22AVLKK61 pKa = 10.43SIEE64 pKa = 4.24NTWPSLYY71 pKa = 10.32KK72 pKa = 10.58YY73 pKa = 10.37EE74 pKa = 4.76IDD76 pKa = 3.57LASKK80 pKa = 9.51VGPMSVRR87 pKa = 11.84KK88 pKa = 8.68PLSEE92 pKa = 3.78RR93 pKa = 11.84MEE95 pKa = 5.37DD96 pKa = 3.3IEE98 pKa = 5.88HH99 pKa = 6.59YY100 pKa = 10.38YY101 pKa = 11.34ADD103 pKa = 3.84ILQSAEE109 pKa = 4.16PLSDD113 pKa = 3.06RR114 pKa = 11.84AIAATIRR121 pKa = 11.84EE122 pKa = 4.28FNRR125 pKa = 11.84LRR127 pKa = 11.84GLRR130 pKa = 11.84LRR132 pKa = 11.84SEE134 pKa = 4.13SRR136 pKa = 11.84TVEE139 pKa = 3.94LMKK142 pKa = 10.75KK143 pKa = 8.14STNSGSPYY151 pKa = 7.45FTKK154 pKa = 10.47RR155 pKa = 11.84RR156 pKa = 11.84LVTDD160 pKa = 3.12KK161 pKa = 10.34TVPSEE166 pKa = 4.31VYY168 pKa = 9.67HH169 pKa = 6.56FRR171 pKa = 11.84DD172 pKa = 3.37EE173 pKa = 4.57VEE175 pKa = 4.02QHH177 pKa = 6.09LSSGDD182 pKa = 3.38FTAAAILGWRR192 pKa = 11.84GQEE195 pKa = 4.13GGPTAADD202 pKa = 3.35VKK204 pKa = 10.98QRR206 pKa = 11.84VVWMFPYY213 pKa = 10.4AVNIAEE219 pKa = 4.25LQLYY223 pKa = 9.45QPFIEE228 pKa = 4.83AAQSSISSQLGLALTRR244 pKa = 11.84LINGSLPCSTQRR256 pKa = 11.84QTNDD260 pKa = 3.22LVVCTDD266 pKa = 3.76FSKK269 pKa = 10.9FDD271 pKa = 3.21QHH273 pKa = 7.11FNAEE277 pKa = 4.07MQAAALRR284 pKa = 11.84ILRR287 pKa = 11.84ACMEE291 pKa = 4.39PSMTSTNWLNDD302 pKa = 3.41VYY304 pKa = 9.95PIKK307 pKa = 11.05YY308 pKa = 7.97MIPLAYY314 pKa = 10.08DD315 pKa = 3.12EE316 pKa = 4.66GKK318 pKa = 10.52VRR320 pKa = 11.84FGKK323 pKa = 9.95HH324 pKa = 4.43GMGSGSGGTNCDD336 pKa = 2.62EE337 pKa = 4.46TLAHH341 pKa = 7.44RR342 pKa = 11.84ALQYY346 pKa = 10.24EE347 pKa = 4.71AAQSSGVKK355 pKa = 10.17LNPNSQCLGDD365 pKa = 5.2DD366 pKa = 3.95GVLTYY371 pKa = 10.34PGITVEE377 pKa = 4.98DD378 pKa = 3.74VVKK381 pKa = 10.39AYY383 pKa = 9.76KK384 pKa = 10.32SHH386 pKa = 5.73GLEE389 pKa = 3.97MNLDD393 pKa = 3.4KK394 pKa = 11.05QYY396 pKa = 11.78ASTQDD401 pKa = 3.2CTYY404 pKa = 10.58LRR406 pKa = 11.84RR407 pKa = 11.84WHH409 pKa = 6.68HH410 pKa = 6.13KK411 pKa = 10.17DD412 pKa = 3.14YY413 pKa = 10.73RR414 pKa = 11.84QDD416 pKa = 4.15GICVGVYY423 pKa = 6.78STCRR427 pKa = 11.84ALGRR431 pKa = 11.84LRR433 pKa = 11.84YY434 pKa = 9.64LEE436 pKa = 5.54RR437 pKa = 11.84YY438 pKa = 8.61QNPKK442 pKa = 9.36YY443 pKa = 9.89WDD445 pKa = 3.4AKK447 pKa = 10.45AVALRR452 pKa = 11.84QLSILEE458 pKa = 4.01NVKK461 pKa = 9.79YY462 pKa = 10.47HH463 pKa = 6.1PLKK466 pKa = 10.33EE467 pKa = 4.09QFVDD471 pKa = 4.52FCMKK475 pKa = 9.73RR476 pKa = 11.84DD477 pKa = 3.6KK478 pKa = 11.17YY479 pKa = 11.06RR480 pKa = 11.84LGLDD484 pKa = 3.01IPHH487 pKa = 6.82FFDD490 pKa = 4.36DD491 pKa = 4.24LRR493 pKa = 11.84AITEE497 pKa = 3.98QKK499 pKa = 9.99IDD501 pKa = 4.34DD502 pKa = 4.07MPDD505 pKa = 2.97FLGYY509 pKa = 9.01TRR511 pKa = 11.84TLQSGGDD518 pKa = 3.53PAGGIEE524 pKa = 3.96NWWIVKK530 pKa = 8.25YY531 pKa = 10.64LKK533 pKa = 10.78SKK535 pKa = 10.67
MM1 pKa = 7.87PKK3 pKa = 10.47NNANKK8 pKa = 10.2RR9 pKa = 11.84SRR11 pKa = 11.84DD12 pKa = 3.55YY13 pKa = 10.54FASSFASSANLRR25 pKa = 11.84AYY27 pKa = 10.02FGSVVKK33 pKa = 10.49GQSEE37 pKa = 5.11VYY39 pKa = 8.01DD40 pKa = 3.67TPFARR45 pKa = 11.84DD46 pKa = 3.35EE47 pKa = 4.27STKK50 pKa = 10.55TLLSKK55 pKa = 10.1WKK57 pKa = 10.22AVLKK61 pKa = 10.43SIEE64 pKa = 4.24NTWPSLYY71 pKa = 10.32KK72 pKa = 10.58YY73 pKa = 10.37EE74 pKa = 4.76IDD76 pKa = 3.57LASKK80 pKa = 9.51VGPMSVRR87 pKa = 11.84KK88 pKa = 8.68PLSEE92 pKa = 3.78RR93 pKa = 11.84MEE95 pKa = 5.37DD96 pKa = 3.3IEE98 pKa = 5.88HH99 pKa = 6.59YY100 pKa = 10.38YY101 pKa = 11.34ADD103 pKa = 3.84ILQSAEE109 pKa = 4.16PLSDD113 pKa = 3.06RR114 pKa = 11.84AIAATIRR121 pKa = 11.84EE122 pKa = 4.28FNRR125 pKa = 11.84LRR127 pKa = 11.84GLRR130 pKa = 11.84LRR132 pKa = 11.84SEE134 pKa = 4.13SRR136 pKa = 11.84TVEE139 pKa = 3.94LMKK142 pKa = 10.75KK143 pKa = 8.14STNSGSPYY151 pKa = 7.45FTKK154 pKa = 10.47RR155 pKa = 11.84RR156 pKa = 11.84LVTDD160 pKa = 3.12KK161 pKa = 10.34TVPSEE166 pKa = 4.31VYY168 pKa = 9.67HH169 pKa = 6.56FRR171 pKa = 11.84DD172 pKa = 3.37EE173 pKa = 4.57VEE175 pKa = 4.02QHH177 pKa = 6.09LSSGDD182 pKa = 3.38FTAAAILGWRR192 pKa = 11.84GQEE195 pKa = 4.13GGPTAADD202 pKa = 3.35VKK204 pKa = 10.98QRR206 pKa = 11.84VVWMFPYY213 pKa = 10.4AVNIAEE219 pKa = 4.25LQLYY223 pKa = 9.45QPFIEE228 pKa = 4.83AAQSSISSQLGLALTRR244 pKa = 11.84LINGSLPCSTQRR256 pKa = 11.84QTNDD260 pKa = 3.22LVVCTDD266 pKa = 3.76FSKK269 pKa = 10.9FDD271 pKa = 3.21QHH273 pKa = 7.11FNAEE277 pKa = 4.07MQAAALRR284 pKa = 11.84ILRR287 pKa = 11.84ACMEE291 pKa = 4.39PSMTSTNWLNDD302 pKa = 3.41VYY304 pKa = 9.95PIKK307 pKa = 11.05YY308 pKa = 7.97MIPLAYY314 pKa = 10.08DD315 pKa = 3.12EE316 pKa = 4.66GKK318 pKa = 10.52VRR320 pKa = 11.84FGKK323 pKa = 9.95HH324 pKa = 4.43GMGSGSGGTNCDD336 pKa = 2.62EE337 pKa = 4.46TLAHH341 pKa = 7.44RR342 pKa = 11.84ALQYY346 pKa = 10.24EE347 pKa = 4.71AAQSSGVKK355 pKa = 10.17LNPNSQCLGDD365 pKa = 5.2DD366 pKa = 3.95GVLTYY371 pKa = 10.34PGITVEE377 pKa = 4.98DD378 pKa = 3.74VVKK381 pKa = 10.39AYY383 pKa = 9.76KK384 pKa = 10.32SHH386 pKa = 5.73GLEE389 pKa = 3.97MNLDD393 pKa = 3.4KK394 pKa = 11.05QYY396 pKa = 11.78ASTQDD401 pKa = 3.2CTYY404 pKa = 10.58LRR406 pKa = 11.84RR407 pKa = 11.84WHH409 pKa = 6.68HH410 pKa = 6.13KK411 pKa = 10.17DD412 pKa = 3.14YY413 pKa = 10.73RR414 pKa = 11.84QDD416 pKa = 4.15GICVGVYY423 pKa = 6.78STCRR427 pKa = 11.84ALGRR431 pKa = 11.84LRR433 pKa = 11.84YY434 pKa = 9.64LEE436 pKa = 5.54RR437 pKa = 11.84YY438 pKa = 8.61QNPKK442 pKa = 9.36YY443 pKa = 9.89WDD445 pKa = 3.4AKK447 pKa = 10.45AVALRR452 pKa = 11.84QLSILEE458 pKa = 4.01NVKK461 pKa = 9.79YY462 pKa = 10.47HH463 pKa = 6.1PLKK466 pKa = 10.33EE467 pKa = 4.09QFVDD471 pKa = 4.52FCMKK475 pKa = 9.73RR476 pKa = 11.84DD477 pKa = 3.6KK478 pKa = 11.17YY479 pKa = 11.06RR480 pKa = 11.84LGLDD484 pKa = 3.01IPHH487 pKa = 6.82FFDD490 pKa = 4.36DD491 pKa = 4.24LRR493 pKa = 11.84AITEE497 pKa = 3.98QKK499 pKa = 9.99IDD501 pKa = 4.34DD502 pKa = 4.07MPDD505 pKa = 2.97FLGYY509 pKa = 9.01TRR511 pKa = 11.84TLQSGGDD518 pKa = 3.53PAGGIEE524 pKa = 3.96NWWIVKK530 pKa = 8.25YY531 pKa = 10.64LKK533 pKa = 10.78SKK535 pKa = 10.67
Molecular weight: 60.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1328 |
178 |
615 |
442.7 |
49.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.605 ± 0.086 | 1.13 ± 0.34 |
5.949 ± 0.599 | 5.723 ± 1.335 |
3.012 ± 0.458 | 6.551 ± 0.232 |
2.108 ± 0.309 | 4.593 ± 0.299 |
5.045 ± 0.897 | 7.605 ± 0.865 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.313 ± 0.682 | 5.12 ± 0.785 |
3.614 ± 0.709 | 4.292 ± 0.957 |
6.325 ± 1.079 | 7.831 ± 0.571 |
6.627 ± 0.773 | 6.702 ± 0.717 |
1.28 ± 0.227 | 5.572 ± 1.222 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
