
Muribaculum sp. An287
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; unclassified Muribaculum
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
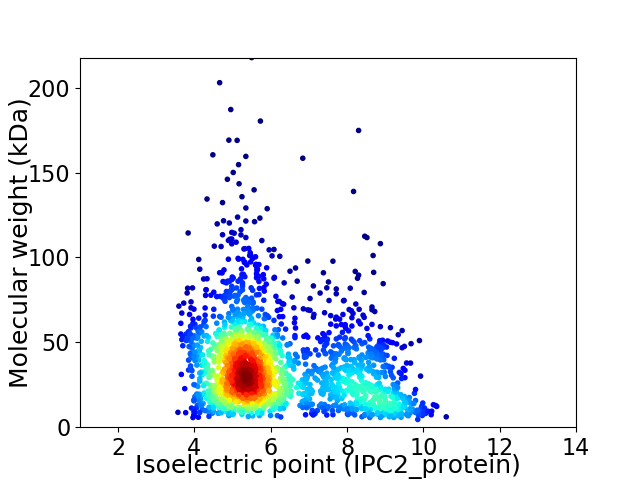
Virtual 2D-PAGE plot for 2021 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4CFU6|A0A1Y4CFU6_9BACT TPM_phosphatase domain-containing protein OS=Muribaculum sp. An287 OX=1965623 GN=B5F81_05730 PE=4 SV=1
MM1 pKa = 7.2KK2 pKa = 10.35RR3 pKa = 11.84ILIFMLSCVSAAAIIGCEE21 pKa = 3.98KK22 pKa = 10.37QIEE25 pKa = 4.48SEE27 pKa = 4.32PPVLSSDD34 pKa = 3.26AKK36 pKa = 10.46DD37 pKa = 3.11IHH39 pKa = 5.69VTAEE43 pKa = 4.26GGTQTITYY51 pKa = 9.54NIEE54 pKa = 3.68NPAEE58 pKa = 4.44DD59 pKa = 4.14GMITALPDD67 pKa = 4.5SEE69 pKa = 5.25CDD71 pKa = 3.22WIKK74 pKa = 9.94ITDD77 pKa = 3.88TEE79 pKa = 4.43TSGVITFNVDD89 pKa = 3.52KK90 pKa = 11.34NEE92 pKa = 3.89TDD94 pKa = 3.29AKK96 pKa = 9.97RR97 pKa = 11.84EE98 pKa = 4.06SKK100 pKa = 9.06ITVTYY105 pKa = 9.14EE106 pKa = 3.72YY107 pKa = 11.06SGGDD111 pKa = 3.41PQSFSVNVSQDD122 pKa = 3.15SGEE125 pKa = 4.34NNPDD129 pKa = 4.17PDD131 pKa = 5.27PDD133 pKa = 5.0PDD135 pKa = 5.1PDD137 pKa = 4.62PDD139 pKa = 4.38PNPGEE144 pKa = 4.22DD145 pKa = 3.26VFTITVDD152 pKa = 3.32NITYY156 pKa = 7.96STAHH160 pKa = 6.22IRR162 pKa = 11.84VVPDD166 pKa = 4.14DD167 pKa = 3.52EE168 pKa = 4.41TMPYY172 pKa = 9.48IVYY175 pKa = 10.47VDD177 pKa = 3.56MASEE181 pKa = 4.15IKK183 pKa = 10.49DD184 pKa = 3.67LSDD187 pKa = 5.03DD188 pKa = 3.87EE189 pKa = 4.67VFQMDD194 pKa = 3.65YY195 pKa = 11.24DD196 pKa = 4.28IIVGNANASQITINEE211 pKa = 4.11YY212 pKa = 10.61LNTIVKK218 pKa = 10.08HH219 pKa = 6.0GPQEE223 pKa = 4.21TTQKK227 pKa = 10.86SLNPGNEE234 pKa = 4.04YY235 pKa = 10.9AVWAYY240 pKa = 10.45GIDD243 pKa = 3.72LQEE246 pKa = 5.41DD247 pKa = 4.04GTLVRR252 pKa = 11.84LSKK255 pKa = 9.49ITRR258 pKa = 11.84EE259 pKa = 4.06YY260 pKa = 9.42FTTEE264 pKa = 3.45IPPVTEE270 pKa = 3.62NSITLEE276 pKa = 4.09VEE278 pKa = 4.2STGTTLSAVATPYY291 pKa = 11.04TNDD294 pKa = 3.31KK295 pKa = 10.77YY296 pKa = 11.98YY297 pKa = 11.04NLNWNNEE304 pKa = 4.16STLVNAGYY312 pKa = 9.14TDD314 pKa = 3.43GTVAEE319 pKa = 4.39RR320 pKa = 11.84CFQFEE325 pKa = 4.13YY326 pKa = 10.7DD327 pKa = 3.9YY328 pKa = 9.9MIKK331 pKa = 10.51YY332 pKa = 9.9ISFYY336 pKa = 11.39AVTDD340 pKa = 3.64WANVKK345 pKa = 10.01QGPATTSTVMTKK357 pKa = 10.28PDD359 pKa = 3.34TYY361 pKa = 11.57YY362 pKa = 11.29LFAYY366 pKa = 8.6FINNDD371 pKa = 2.78ATADD375 pKa = 3.7GEE377 pKa = 4.64IILKK381 pKa = 9.33TIEE384 pKa = 3.75FDD386 pKa = 3.53GANVTIRR393 pKa = 3.51
MM1 pKa = 7.2KK2 pKa = 10.35RR3 pKa = 11.84ILIFMLSCVSAAAIIGCEE21 pKa = 3.98KK22 pKa = 10.37QIEE25 pKa = 4.48SEE27 pKa = 4.32PPVLSSDD34 pKa = 3.26AKK36 pKa = 10.46DD37 pKa = 3.11IHH39 pKa = 5.69VTAEE43 pKa = 4.26GGTQTITYY51 pKa = 9.54NIEE54 pKa = 3.68NPAEE58 pKa = 4.44DD59 pKa = 4.14GMITALPDD67 pKa = 4.5SEE69 pKa = 5.25CDD71 pKa = 3.22WIKK74 pKa = 9.94ITDD77 pKa = 3.88TEE79 pKa = 4.43TSGVITFNVDD89 pKa = 3.52KK90 pKa = 11.34NEE92 pKa = 3.89TDD94 pKa = 3.29AKK96 pKa = 9.97RR97 pKa = 11.84EE98 pKa = 4.06SKK100 pKa = 9.06ITVTYY105 pKa = 9.14EE106 pKa = 3.72YY107 pKa = 11.06SGGDD111 pKa = 3.41PQSFSVNVSQDD122 pKa = 3.15SGEE125 pKa = 4.34NNPDD129 pKa = 4.17PDD131 pKa = 5.27PDD133 pKa = 5.0PDD135 pKa = 5.1PDD137 pKa = 4.62PDD139 pKa = 4.38PNPGEE144 pKa = 4.22DD145 pKa = 3.26VFTITVDD152 pKa = 3.32NITYY156 pKa = 7.96STAHH160 pKa = 6.22IRR162 pKa = 11.84VVPDD166 pKa = 4.14DD167 pKa = 3.52EE168 pKa = 4.41TMPYY172 pKa = 9.48IVYY175 pKa = 10.47VDD177 pKa = 3.56MASEE181 pKa = 4.15IKK183 pKa = 10.49DD184 pKa = 3.67LSDD187 pKa = 5.03DD188 pKa = 3.87EE189 pKa = 4.67VFQMDD194 pKa = 3.65YY195 pKa = 11.24DD196 pKa = 4.28IIVGNANASQITINEE211 pKa = 4.11YY212 pKa = 10.61LNTIVKK218 pKa = 10.08HH219 pKa = 6.0GPQEE223 pKa = 4.21TTQKK227 pKa = 10.86SLNPGNEE234 pKa = 4.04YY235 pKa = 10.9AVWAYY240 pKa = 10.45GIDD243 pKa = 3.72LQEE246 pKa = 5.41DD247 pKa = 4.04GTLVRR252 pKa = 11.84LSKK255 pKa = 9.49ITRR258 pKa = 11.84EE259 pKa = 4.06YY260 pKa = 9.42FTTEE264 pKa = 3.45IPPVTEE270 pKa = 3.62NSITLEE276 pKa = 4.09VEE278 pKa = 4.2STGTTLSAVATPYY291 pKa = 11.04TNDD294 pKa = 3.31KK295 pKa = 10.77YY296 pKa = 11.98YY297 pKa = 11.04NLNWNNEE304 pKa = 4.16STLVNAGYY312 pKa = 9.14TDD314 pKa = 3.43GTVAEE319 pKa = 4.39RR320 pKa = 11.84CFQFEE325 pKa = 4.13YY326 pKa = 10.7DD327 pKa = 3.9YY328 pKa = 9.9MIKK331 pKa = 10.51YY332 pKa = 9.9ISFYY336 pKa = 11.39AVTDD340 pKa = 3.64WANVKK345 pKa = 10.01QGPATTSTVMTKK357 pKa = 10.28PDD359 pKa = 3.34TYY361 pKa = 11.57YY362 pKa = 11.29LFAYY366 pKa = 8.6FINNDD371 pKa = 2.78ATADD375 pKa = 3.7GEE377 pKa = 4.64IILKK381 pKa = 9.33TIEE384 pKa = 3.75FDD386 pKa = 3.53GANVTIRR393 pKa = 3.51
Molecular weight: 43.64 kDa
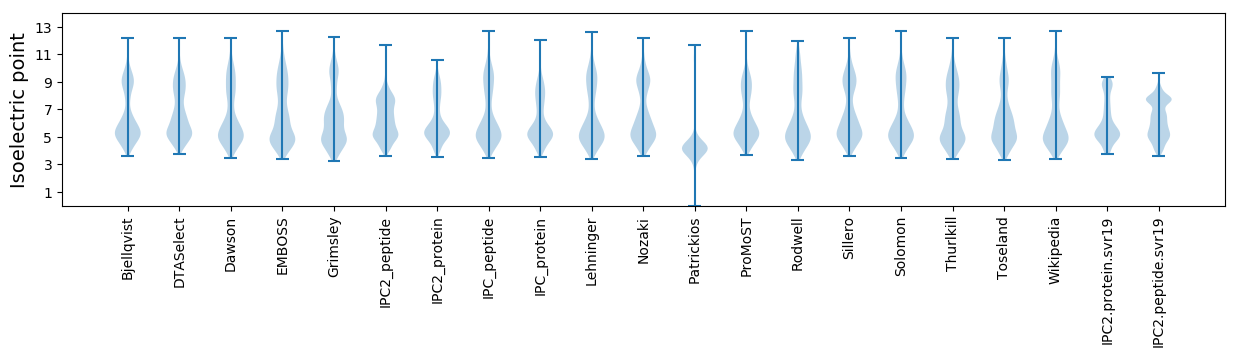
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4CL05|A0A1Y4CL05_9BACT Uncharacterized protein OS=Muribaculum sp. An287 OX=1965623 GN=B5F81_02195 PE=4 SV=1
MM1 pKa = 7.78EE2 pKa = 4.06YY3 pKa = 10.44SINKK7 pKa = 9.56GIGRR11 pKa = 11.84SVEE14 pKa = 4.22FKK16 pKa = 10.81GLKK19 pKa = 9.21AQYY22 pKa = 11.01LFIFCGGLLAVFVLFMVMYY41 pKa = 8.69MAGVAQGVCVGFGAVSASALIWLTFRR67 pKa = 11.84LNARR71 pKa = 11.84YY72 pKa = 9.82GEE74 pKa = 4.32HH75 pKa = 7.16GLMKK79 pKa = 10.59LGALRR84 pKa = 11.84SRR86 pKa = 11.84PRR88 pKa = 11.84YY89 pKa = 8.8IINRR93 pKa = 11.84RR94 pKa = 11.84AVFRR98 pKa = 11.84LFKK101 pKa = 9.99QTDD104 pKa = 3.19RR105 pKa = 11.84KK106 pKa = 9.94EE107 pKa = 4.35AIRR110 pKa = 4.45
MM1 pKa = 7.78EE2 pKa = 4.06YY3 pKa = 10.44SINKK7 pKa = 9.56GIGRR11 pKa = 11.84SVEE14 pKa = 4.22FKK16 pKa = 10.81GLKK19 pKa = 9.21AQYY22 pKa = 11.01LFIFCGGLLAVFVLFMVMYY41 pKa = 8.69MAGVAQGVCVGFGAVSASALIWLTFRR67 pKa = 11.84LNARR71 pKa = 11.84YY72 pKa = 9.82GEE74 pKa = 4.32HH75 pKa = 7.16GLMKK79 pKa = 10.59LGALRR84 pKa = 11.84SRR86 pKa = 11.84PRR88 pKa = 11.84YY89 pKa = 8.8IINRR93 pKa = 11.84RR94 pKa = 11.84AVFRR98 pKa = 11.84LFKK101 pKa = 9.99QTDD104 pKa = 3.19RR105 pKa = 11.84KK106 pKa = 9.94EE107 pKa = 4.35AIRR110 pKa = 4.45
Molecular weight: 12.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
695823 |
38 |
1912 |
344.3 |
38.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.922 ± 0.056 | 1.332 ± 0.021 |
5.978 ± 0.037 | 6.887 ± 0.054 |
4.566 ± 0.037 | 7.661 ± 0.044 |
1.611 ± 0.024 | 6.947 ± 0.051 |
5.858 ± 0.049 | 8.554 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.887 ± 0.027 | 4.267 ± 0.036 |
3.809 ± 0.032 | 2.519 ± 0.03 |
5.202 ± 0.046 | 6.744 ± 0.048 |
5.187 ± 0.042 | 6.783 ± 0.041 |
1.066 ± 0.02 | 4.22 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
