
Sewage-associated circular DNA virus-31
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
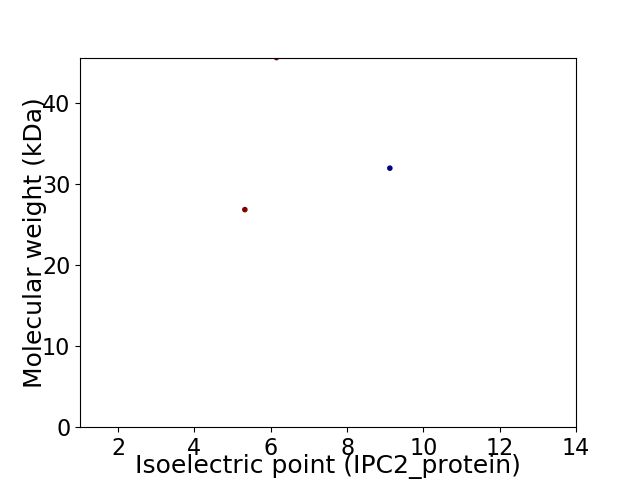
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
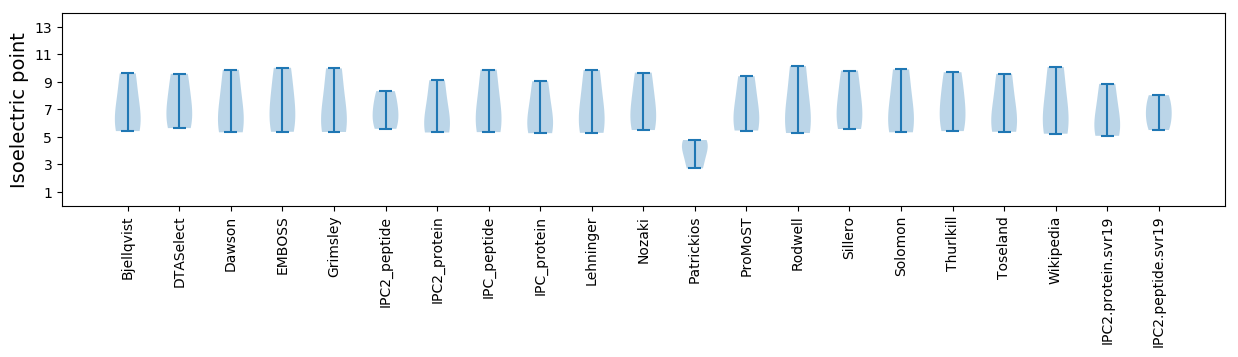
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGZ8|A0A0B4UGZ8_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-31 OX=1592098 PE=4 SV=1
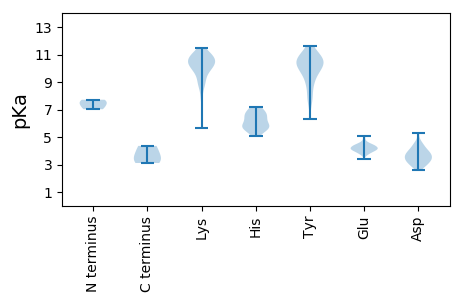
MM1 pKa = 7.71PARR4 pKa = 11.84LEE6 pKa = 3.99IEE8 pKa = 4.53SYY10 pKa = 10.73SSYY13 pKa = 11.05LIYY16 pKa = 11.03LNLFLSIYY24 pKa = 8.95FLKK27 pKa = 10.94YY28 pKa = 7.92QTNLMAYY35 pKa = 8.95KK36 pKa = 10.68DD37 pKa = 3.91SANTANKK44 pKa = 10.07YY45 pKa = 10.32FLLLGDD51 pKa = 5.31RR52 pKa = 11.84ILDD55 pKa = 3.79CVEE58 pKa = 4.41LNPDD62 pKa = 4.26GVCEE66 pKa = 4.2ADD68 pKa = 4.56DD69 pKa = 3.57VQLFINRR76 pKa = 11.84SSIFLSDD83 pKa = 3.55VGLKK87 pKa = 8.78AHH89 pKa = 6.61KK90 pKa = 9.28WVIGQLHH97 pKa = 7.19HH98 pKa = 6.95YY99 pKa = 9.31NNEE102 pKa = 3.94YY103 pKa = 9.75TKK105 pKa = 11.24GRR107 pKa = 11.84MSEE110 pKa = 4.11HH111 pKa = 6.57EE112 pKa = 4.12LVEE115 pKa = 4.36AYY117 pKa = 10.26GYY119 pKa = 11.32LNLEE123 pKa = 4.17GYY125 pKa = 9.22LRR127 pKa = 11.84HH128 pKa = 5.85VAYY131 pKa = 7.6EE132 pKa = 4.09TYY134 pKa = 10.66KK135 pKa = 10.26EE136 pKa = 4.16VRR138 pKa = 11.84GKK140 pKa = 9.52WEE142 pKa = 4.07TRR144 pKa = 11.84VVEE147 pKa = 4.07YY148 pKa = 10.69DD149 pKa = 3.41LKK151 pKa = 11.49GDD153 pKa = 3.77VASVNYY159 pKa = 9.78CLRR162 pKa = 11.84RR163 pKa = 11.84LEE165 pKa = 4.21LMAIARR171 pKa = 11.84SQGTTLKK178 pKa = 10.78QIFKK182 pKa = 9.72SNGWEE187 pKa = 4.46GYY189 pKa = 10.18LDD191 pKa = 4.54RR192 pKa = 11.84EE193 pKa = 4.36YY194 pKa = 11.22QPIEE198 pKa = 3.95YY199 pKa = 10.35QFVDD203 pKa = 4.73PYY205 pKa = 11.6LLDD208 pKa = 3.58KK209 pKa = 10.78FGRR212 pKa = 11.84EE213 pKa = 3.83AWEE216 pKa = 4.1YY217 pKa = 10.54TPDD220 pKa = 3.7PYY222 pKa = 10.68ATHH225 pKa = 6.91KK226 pKa = 11.03GSLL229 pKa = 3.55
MM1 pKa = 7.71PARR4 pKa = 11.84LEE6 pKa = 3.99IEE8 pKa = 4.53SYY10 pKa = 10.73SSYY13 pKa = 11.05LIYY16 pKa = 11.03LNLFLSIYY24 pKa = 8.95FLKK27 pKa = 10.94YY28 pKa = 7.92QTNLMAYY35 pKa = 8.95KK36 pKa = 10.68DD37 pKa = 3.91SANTANKK44 pKa = 10.07YY45 pKa = 10.32FLLLGDD51 pKa = 5.31RR52 pKa = 11.84ILDD55 pKa = 3.79CVEE58 pKa = 4.41LNPDD62 pKa = 4.26GVCEE66 pKa = 4.2ADD68 pKa = 4.56DD69 pKa = 3.57VQLFINRR76 pKa = 11.84SSIFLSDD83 pKa = 3.55VGLKK87 pKa = 8.78AHH89 pKa = 6.61KK90 pKa = 9.28WVIGQLHH97 pKa = 7.19HH98 pKa = 6.95YY99 pKa = 9.31NNEE102 pKa = 3.94YY103 pKa = 9.75TKK105 pKa = 11.24GRR107 pKa = 11.84MSEE110 pKa = 4.11HH111 pKa = 6.57EE112 pKa = 4.12LVEE115 pKa = 4.36AYY117 pKa = 10.26GYY119 pKa = 11.32LNLEE123 pKa = 4.17GYY125 pKa = 9.22LRR127 pKa = 11.84HH128 pKa = 5.85VAYY131 pKa = 7.6EE132 pKa = 4.09TYY134 pKa = 10.66KK135 pKa = 10.26EE136 pKa = 4.16VRR138 pKa = 11.84GKK140 pKa = 9.52WEE142 pKa = 4.07TRR144 pKa = 11.84VVEE147 pKa = 4.07YY148 pKa = 10.69DD149 pKa = 3.41LKK151 pKa = 11.49GDD153 pKa = 3.77VASVNYY159 pKa = 9.78CLRR162 pKa = 11.84RR163 pKa = 11.84LEE165 pKa = 4.21LMAIARR171 pKa = 11.84SQGTTLKK178 pKa = 10.78QIFKK182 pKa = 9.72SNGWEE187 pKa = 4.46GYY189 pKa = 10.18LDD191 pKa = 4.54RR192 pKa = 11.84EE193 pKa = 4.36YY194 pKa = 11.22QPIEE198 pKa = 3.95YY199 pKa = 10.35QFVDD203 pKa = 4.73PYY205 pKa = 11.6LLDD208 pKa = 3.58KK209 pKa = 10.78FGRR212 pKa = 11.84EE213 pKa = 3.83AWEE216 pKa = 4.1YY217 pKa = 10.54TPDD220 pKa = 3.7PYY222 pKa = 10.68ATHH225 pKa = 6.91KK226 pKa = 11.03GSLL229 pKa = 3.55
Molecular weight: 26.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGN7|A0A0B4UGN7_9VIRU Uncharacterized protein OS=Sewage-associated circular DNA virus-31 OX=1592098 PE=4 SV=1
MM1 pKa = 7.02YY2 pKa = 10.59GKK4 pKa = 10.43FRR6 pKa = 11.84GSKK9 pKa = 8.75GASSRR14 pKa = 11.84RR15 pKa = 11.84TNGITANNIARR26 pKa = 11.84ALKK29 pKa = 10.35KK30 pKa = 10.84VGGKK34 pKa = 7.84YY35 pKa = 8.26TSKK38 pKa = 10.53KK39 pKa = 9.05YY40 pKa = 9.72SAPRR44 pKa = 11.84FATVGFARR52 pKa = 11.84NVEE55 pKa = 3.97KK56 pKa = 10.62KK57 pKa = 10.6YY58 pKa = 10.77FDD60 pKa = 3.2KK61 pKa = 11.11QYY63 pKa = 10.94ISNSLEE69 pKa = 3.73ALTGITGNSASNGVTYY85 pKa = 10.62ISNVWGSYY93 pKa = 9.84NFGAQVASTAISNDD107 pKa = 2.82MCKK110 pKa = 10.48GLDD113 pKa = 3.2TGTTARR119 pKa = 11.84TRR121 pKa = 11.84IGNKK125 pKa = 9.97LKK127 pKa = 10.68VSYY130 pKa = 10.45IKK132 pKa = 10.9GAFTFTAAQSNGMNNTPQNGEE153 pKa = 4.0NQVVPLAANTKK164 pKa = 8.97QDD166 pKa = 3.87YY167 pKa = 11.12LRR169 pKa = 11.84TTYY172 pKa = 10.81RR173 pKa = 11.84FVIVKK178 pKa = 9.64DD179 pKa = 3.6LQVNSTDD186 pKa = 4.11AQVTWAQVFDD196 pKa = 4.07TTGSQAGVHH205 pKa = 6.45AEE207 pKa = 4.0LNVDD211 pKa = 2.87NMGRR215 pKa = 11.84FIVIQDD221 pKa = 3.41KK222 pKa = 11.18YY223 pKa = 10.28FTLDD227 pKa = 3.16AQNPQQTCPFQVKK240 pKa = 9.91GSSVGNVRR248 pKa = 11.84YY249 pKa = 9.87NGPGAAALTDD259 pKa = 3.24KK260 pKa = 11.09GIYY263 pKa = 8.44VVWAAYY269 pKa = 10.43VMGVDD274 pKa = 4.37GVGTANVQCPSPVGHH289 pKa = 6.58SRR291 pKa = 11.84LCFTDD296 pKa = 3.9DD297 pKa = 3.11
MM1 pKa = 7.02YY2 pKa = 10.59GKK4 pKa = 10.43FRR6 pKa = 11.84GSKK9 pKa = 8.75GASSRR14 pKa = 11.84RR15 pKa = 11.84TNGITANNIARR26 pKa = 11.84ALKK29 pKa = 10.35KK30 pKa = 10.84VGGKK34 pKa = 7.84YY35 pKa = 8.26TSKK38 pKa = 10.53KK39 pKa = 9.05YY40 pKa = 9.72SAPRR44 pKa = 11.84FATVGFARR52 pKa = 11.84NVEE55 pKa = 3.97KK56 pKa = 10.62KK57 pKa = 10.6YY58 pKa = 10.77FDD60 pKa = 3.2KK61 pKa = 11.11QYY63 pKa = 10.94ISNSLEE69 pKa = 3.73ALTGITGNSASNGVTYY85 pKa = 10.62ISNVWGSYY93 pKa = 9.84NFGAQVASTAISNDD107 pKa = 2.82MCKK110 pKa = 10.48GLDD113 pKa = 3.2TGTTARR119 pKa = 11.84TRR121 pKa = 11.84IGNKK125 pKa = 9.97LKK127 pKa = 10.68VSYY130 pKa = 10.45IKK132 pKa = 10.9GAFTFTAAQSNGMNNTPQNGEE153 pKa = 4.0NQVVPLAANTKK164 pKa = 8.97QDD166 pKa = 3.87YY167 pKa = 11.12LRR169 pKa = 11.84TTYY172 pKa = 10.81RR173 pKa = 11.84FVIVKK178 pKa = 9.64DD179 pKa = 3.6LQVNSTDD186 pKa = 4.11AQVTWAQVFDD196 pKa = 4.07TTGSQAGVHH205 pKa = 6.45AEE207 pKa = 4.0LNVDD211 pKa = 2.87NMGRR215 pKa = 11.84FIVIQDD221 pKa = 3.41KK222 pKa = 11.18YY223 pKa = 10.28FTLDD227 pKa = 3.16AQNPQQTCPFQVKK240 pKa = 9.91GSSVGNVRR248 pKa = 11.84YY249 pKa = 9.87NGPGAAALTDD259 pKa = 3.24KK260 pKa = 11.09GIYY263 pKa = 8.44VVWAAYY269 pKa = 10.43VMGVDD274 pKa = 4.37GVGTANVQCPSPVGHH289 pKa = 6.58SRR291 pKa = 11.84LCFTDD296 pKa = 3.9DD297 pKa = 3.11
Molecular weight: 32.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
924 |
229 |
398 |
308.0 |
34.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.359 ± 0.979 | 1.84 ± 0.352 |
5.087 ± 0.205 | 6.277 ± 2.007 |
3.896 ± 0.2 | 8.658 ± 0.861 |
1.84 ± 0.473 | 4.437 ± 0.192 |
6.818 ± 0.526 | 6.385 ± 2.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.056 ± 0.24 | 5.736 ± 0.956 |
3.03 ± 0.255 | 4.437 ± 0.493 |
5.087 ± 0.299 | 5.303 ± 0.865 |
6.602 ± 1.109 | 7.143 ± 0.799 |
2.381 ± 0.749 | 5.628 ± 1.313 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
