
Bat coronavirus HKU9 (BtCoV) (BtCoV/HKU9)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Nobecovirus
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
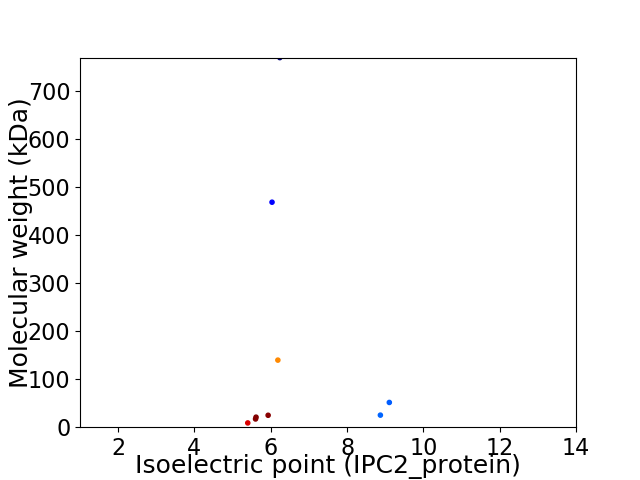
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
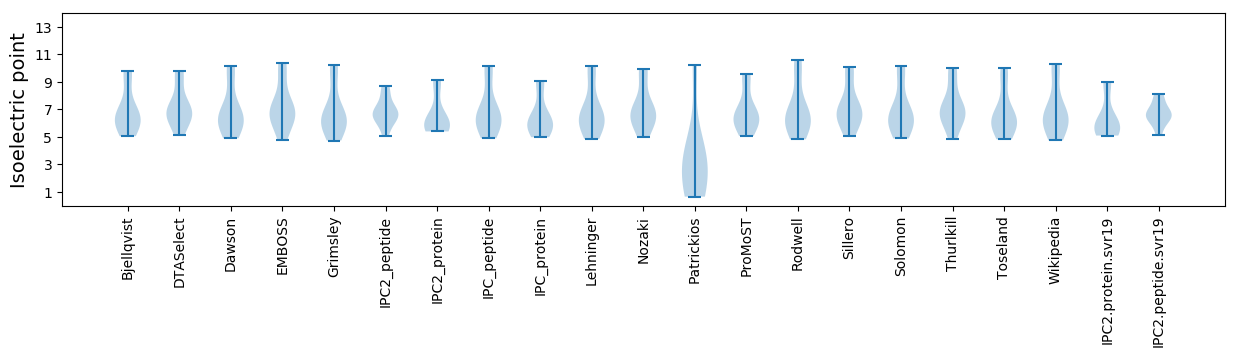
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A3EXG9|VME1_BCHK9 Membrane protein OS=Bat coronavirus HKU9 OX=694006 GN=M PE=3 SV=1
MM1 pKa = 7.31YY2 pKa = 10.07DD3 pKa = 3.06IVGTNNSILIANVLVLIIICLLVVIVGCALLLILQFVFGVCGFVFKK49 pKa = 9.25FVCKK53 pKa = 8.8PTILVYY59 pKa = 11.02NKK61 pKa = 9.42FRR63 pKa = 11.84NEE65 pKa = 3.56SLLNEE70 pKa = 4.19RR71 pKa = 11.84EE72 pKa = 4.03EE73 pKa = 4.17LLCDD77 pKa = 3.64NVV79 pKa = 3.42
MM1 pKa = 7.31YY2 pKa = 10.07DD3 pKa = 3.06IVGTNNSILIANVLVLIIICLLVVIVGCALLLILQFVFGVCGFVFKK49 pKa = 9.25FVCKK53 pKa = 8.8PTILVYY59 pKa = 11.02NKK61 pKa = 9.42FRR63 pKa = 11.84NEE65 pKa = 3.56SLLNEE70 pKa = 4.19RR71 pKa = 11.84EE72 pKa = 4.03EE73 pKa = 4.17LLCDD77 pKa = 3.64NVV79 pKa = 3.42
Molecular weight: 8.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A3EXH1|NS7A_BCHK9 Non-structural protein 7a OS=Bat coronavirus HKU9 OX=694006 GN=7a PE=3 SV=1
MM1 pKa = 7.53SGRR4 pKa = 11.84NRR6 pKa = 11.84SRR8 pKa = 11.84SGTPSPKK15 pKa = 9.14VTFKK19 pKa = 11.02QEE21 pKa = 3.89SDD23 pKa = 3.52GSDD26 pKa = 3.37SEE28 pKa = 4.52SEE30 pKa = 3.92RR31 pKa = 11.84RR32 pKa = 11.84NGNRR36 pKa = 11.84NGARR40 pKa = 11.84PKK42 pKa = 10.5NNNSRR47 pKa = 11.84GSAPKK52 pKa = 9.89PEE54 pKa = 4.43KK55 pKa = 10.23PKK57 pKa = 10.56AAPPQNVSWFAPLVQTGKK75 pKa = 10.94AEE77 pKa = 3.93LRR79 pKa = 11.84FPRR82 pKa = 11.84GEE84 pKa = 4.09GVPVSQGVDD93 pKa = 2.91STYY96 pKa = 11.3EE97 pKa = 3.4HH98 pKa = 7.18GYY100 pKa = 8.24WLRR103 pKa = 11.84TQRR106 pKa = 11.84SFQKK110 pKa = 10.63GGKK113 pKa = 8.51QVLANPRR120 pKa = 11.84WYY122 pKa = 10.14FYY124 pKa = 10.09YY125 pKa = 10.29TGTGRR130 pKa = 11.84FGDD133 pKa = 3.82LRR135 pKa = 11.84FGTKK139 pKa = 10.25NPDD142 pKa = 3.36IVWVGQEE149 pKa = 3.73GANINRR155 pKa = 11.84LGDD158 pKa = 3.28MGTRR162 pKa = 11.84NPSNDD167 pKa = 2.83GAIPVQLAGGIPKK180 pKa = 10.3GFYY183 pKa = 10.56AEE185 pKa = 4.4GRR187 pKa = 11.84GSRR190 pKa = 11.84GNSRR194 pKa = 11.84SSSRR198 pKa = 11.84NSSRR202 pKa = 11.84ASSRR206 pKa = 11.84GNSRR210 pKa = 11.84ASSRR214 pKa = 11.84GASPGRR220 pKa = 11.84PAANPSTEE228 pKa = 3.28PWMAYY233 pKa = 9.55LVQKK237 pKa = 10.22LEE239 pKa = 4.11RR240 pKa = 11.84LEE242 pKa = 4.27SQVSGTKK249 pKa = 9.77PATKK253 pKa = 10.5NPVQVTKK260 pKa = 11.33NEE262 pKa = 3.71AAANAKK268 pKa = 9.82KK269 pKa = 10.2LRR271 pKa = 11.84HH272 pKa = 6.07KK273 pKa = 9.87RR274 pKa = 11.84TAHH277 pKa = 6.37KK278 pKa = 10.71GSGVTVNYY286 pKa = 9.67GRR288 pKa = 11.84RR289 pKa = 11.84GPGDD293 pKa = 3.4LEE295 pKa = 4.89GNFGDD300 pKa = 4.8RR301 pKa = 11.84EE302 pKa = 4.21MIKK305 pKa = 10.58LGTDD309 pKa = 3.39DD310 pKa = 4.27PRR312 pKa = 11.84FAAAAQMAPNVSSFLFMSHH331 pKa = 7.13LSTRR335 pKa = 11.84DD336 pKa = 3.01EE337 pKa = 5.85DD338 pKa = 3.98DD339 pKa = 5.0ALWLHH344 pKa = 5.98YY345 pKa = 10.2KK346 pKa = 10.54GAIKK350 pKa = 10.4LPKK353 pKa = 9.74DD354 pKa = 3.61DD355 pKa = 5.0PNYY358 pKa = 9.53EE359 pKa = 3.75QWTKK363 pKa = 10.82ILAEE367 pKa = 4.18NLNAYY372 pKa = 9.73KK373 pKa = 10.34DD374 pKa = 4.19FPPTEE379 pKa = 4.01PKK381 pKa = 9.85KK382 pKa = 10.54DD383 pKa = 3.37KK384 pKa = 10.64KK385 pKa = 10.79KK386 pKa = 10.82KK387 pKa = 10.43EE388 pKa = 3.69EE389 pKa = 3.97TAQDD393 pKa = 3.47TVIFEE398 pKa = 4.52DD399 pKa = 4.13ASTGTDD405 pKa = 3.18QTVVKK410 pKa = 10.27VWVKK414 pKa = 10.92DD415 pKa = 3.4QDD417 pKa = 3.61AQTDD421 pKa = 4.02DD422 pKa = 3.18EE423 pKa = 4.56WLGGDD428 pKa = 3.28EE429 pKa = 4.37TVYY432 pKa = 10.85EE433 pKa = 4.85DD434 pKa = 4.34EE435 pKa = 5.21DD436 pKa = 4.66DD437 pKa = 3.79RR438 pKa = 11.84PKK440 pKa = 9.49TQRR443 pKa = 11.84RR444 pKa = 11.84HH445 pKa = 5.22KK446 pKa = 10.5KK447 pKa = 9.72RR448 pKa = 11.84GSTASRR454 pKa = 11.84VTIADD459 pKa = 3.46PTNAGAEE466 pKa = 4.13RR467 pKa = 11.84SS468 pKa = 3.74
MM1 pKa = 7.53SGRR4 pKa = 11.84NRR6 pKa = 11.84SRR8 pKa = 11.84SGTPSPKK15 pKa = 9.14VTFKK19 pKa = 11.02QEE21 pKa = 3.89SDD23 pKa = 3.52GSDD26 pKa = 3.37SEE28 pKa = 4.52SEE30 pKa = 3.92RR31 pKa = 11.84RR32 pKa = 11.84NGNRR36 pKa = 11.84NGARR40 pKa = 11.84PKK42 pKa = 10.5NNNSRR47 pKa = 11.84GSAPKK52 pKa = 9.89PEE54 pKa = 4.43KK55 pKa = 10.23PKK57 pKa = 10.56AAPPQNVSWFAPLVQTGKK75 pKa = 10.94AEE77 pKa = 3.93LRR79 pKa = 11.84FPRR82 pKa = 11.84GEE84 pKa = 4.09GVPVSQGVDD93 pKa = 2.91STYY96 pKa = 11.3EE97 pKa = 3.4HH98 pKa = 7.18GYY100 pKa = 8.24WLRR103 pKa = 11.84TQRR106 pKa = 11.84SFQKK110 pKa = 10.63GGKK113 pKa = 8.51QVLANPRR120 pKa = 11.84WYY122 pKa = 10.14FYY124 pKa = 10.09YY125 pKa = 10.29TGTGRR130 pKa = 11.84FGDD133 pKa = 3.82LRR135 pKa = 11.84FGTKK139 pKa = 10.25NPDD142 pKa = 3.36IVWVGQEE149 pKa = 3.73GANINRR155 pKa = 11.84LGDD158 pKa = 3.28MGTRR162 pKa = 11.84NPSNDD167 pKa = 2.83GAIPVQLAGGIPKK180 pKa = 10.3GFYY183 pKa = 10.56AEE185 pKa = 4.4GRR187 pKa = 11.84GSRR190 pKa = 11.84GNSRR194 pKa = 11.84SSSRR198 pKa = 11.84NSSRR202 pKa = 11.84ASSRR206 pKa = 11.84GNSRR210 pKa = 11.84ASSRR214 pKa = 11.84GASPGRR220 pKa = 11.84PAANPSTEE228 pKa = 3.28PWMAYY233 pKa = 9.55LVQKK237 pKa = 10.22LEE239 pKa = 4.11RR240 pKa = 11.84LEE242 pKa = 4.27SQVSGTKK249 pKa = 9.77PATKK253 pKa = 10.5NPVQVTKK260 pKa = 11.33NEE262 pKa = 3.71AAANAKK268 pKa = 9.82KK269 pKa = 10.2LRR271 pKa = 11.84HH272 pKa = 6.07KK273 pKa = 9.87RR274 pKa = 11.84TAHH277 pKa = 6.37KK278 pKa = 10.71GSGVTVNYY286 pKa = 9.67GRR288 pKa = 11.84RR289 pKa = 11.84GPGDD293 pKa = 3.4LEE295 pKa = 4.89GNFGDD300 pKa = 4.8RR301 pKa = 11.84EE302 pKa = 4.21MIKK305 pKa = 10.58LGTDD309 pKa = 3.39DD310 pKa = 4.27PRR312 pKa = 11.84FAAAAQMAPNVSSFLFMSHH331 pKa = 7.13LSTRR335 pKa = 11.84DD336 pKa = 3.01EE337 pKa = 5.85DD338 pKa = 3.98DD339 pKa = 5.0ALWLHH344 pKa = 5.98YY345 pKa = 10.2KK346 pKa = 10.54GAIKK350 pKa = 10.4LPKK353 pKa = 9.74DD354 pKa = 3.61DD355 pKa = 5.0PNYY358 pKa = 9.53EE359 pKa = 3.75QWTKK363 pKa = 10.82ILAEE367 pKa = 4.18NLNAYY372 pKa = 9.73KK373 pKa = 10.34DD374 pKa = 4.19FPPTEE379 pKa = 4.01PKK381 pKa = 9.85KK382 pKa = 10.54DD383 pKa = 3.37KK384 pKa = 10.64KK385 pKa = 10.79KK386 pKa = 10.82KK387 pKa = 10.43EE388 pKa = 3.69EE389 pKa = 3.97TAQDD393 pKa = 3.47TVIFEE398 pKa = 4.52DD399 pKa = 4.13ASTGTDD405 pKa = 3.18QTVVKK410 pKa = 10.27VWVKK414 pKa = 10.92DD415 pKa = 3.4QDD417 pKa = 3.61AQTDD421 pKa = 4.02DD422 pKa = 3.18EE423 pKa = 4.56WLGGDD428 pKa = 3.28EE429 pKa = 4.37TVYY432 pKa = 10.85EE433 pKa = 4.85DD434 pKa = 4.34EE435 pKa = 5.21DD436 pKa = 4.66DD437 pKa = 3.79RR438 pKa = 11.84PKK440 pKa = 9.49TQRR443 pKa = 11.84RR444 pKa = 11.84HH445 pKa = 5.22KK446 pKa = 10.5KK447 pKa = 9.72RR448 pKa = 11.84GSTASRR454 pKa = 11.84VTIADD459 pKa = 3.46PTNAGAEE466 pKa = 4.13RR467 pKa = 11.84SS468 pKa = 3.74
Molecular weight: 51.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13775 |
79 |
6930 |
1530.6 |
169.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.535 ± 0.145 | 3.114 ± 0.289 |
5.314 ± 0.183 | 3.652 ± 0.239 |
4.436 ± 0.25 | 6.475 ± 0.252 |
1.808 ± 0.133 | 4.414 ± 0.34 |
4.53 ± 0.371 | 9.829 ± 0.441 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.505 ± 0.133 | 4.82 ± 0.586 |
4.668 ± 0.258 | 3.071 ± 0.167 |
4.247 ± 0.463 | 6.904 ± 0.398 |
6.628 ± 0.169 | 9.851 ± 0.624 |
1.299 ± 0.109 | 4.9 ± 0.377 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
