
Vibrio phage VfO4K68
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Versovirus; Vibrio virus VfO3K6
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
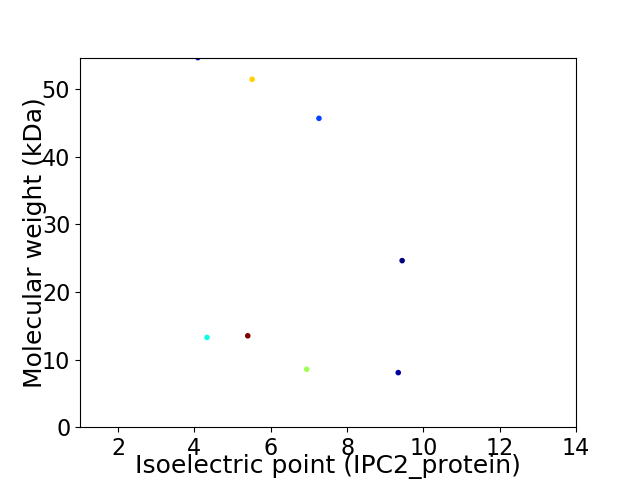
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9MBX5|Q9MBX5_9VIRU Vpf81 OS=Vibrio phage VfO4K68 OX=127508 GN=vpf81 PE=4 SV=1
MM1 pKa = 7.28NKK3 pKa = 10.06SLFLLLFSCLFLSLNASAAQPTYY26 pKa = 10.54KK27 pKa = 10.5VSDD30 pKa = 3.43VSAYY34 pKa = 9.5PDD36 pKa = 3.59CKK38 pKa = 10.69LLLGLRR44 pKa = 11.84VNPASYY50 pKa = 10.56VSCYY54 pKa = 8.61EE55 pKa = 4.05NKK57 pKa = 9.73FVNYY61 pKa = 9.99KK62 pKa = 10.62DD63 pKa = 4.74FSTKK67 pKa = 9.71SCYY70 pKa = 9.75LRR72 pKa = 11.84HH73 pKa = 5.84GKK75 pKa = 9.97YY76 pKa = 9.97VVDD79 pKa = 4.27IMCHH83 pKa = 4.35TTSPSWPLYY92 pKa = 9.22RR93 pKa = 11.84AAGFLQNSAQCPPDD107 pKa = 3.53YY108 pKa = 11.08EE109 pKa = 4.4KK110 pKa = 11.45VEE112 pKa = 4.15DD113 pKa = 4.48EE114 pKa = 4.63YY115 pKa = 11.89VVSCEE120 pKa = 4.44PIVPACEE127 pKa = 3.92YY128 pKa = 11.15GEE130 pKa = 4.53NPDD133 pKa = 4.02GSCMDD138 pKa = 3.64ACQFKK143 pKa = 10.73QSIGDD148 pKa = 3.7TVKK151 pKa = 10.86LYY153 pKa = 8.08WHH155 pKa = 6.48PAIYY159 pKa = 10.72GEE161 pKa = 4.73LVTGACYY168 pKa = 10.65GDD170 pKa = 4.13YY171 pKa = 10.89GATRR175 pKa = 11.84CEE177 pKa = 4.02VTKK180 pKa = 11.03NEE182 pKa = 4.01STIICTGVPDD192 pKa = 4.14GQYY195 pKa = 10.09TPDD198 pKa = 3.67SQCSLRR204 pKa = 11.84FAYY207 pKa = 9.17TGRR210 pKa = 11.84QCDD213 pKa = 3.3GGTLFWGVNGPDD225 pKa = 3.77EE226 pKa = 5.2PIIPPDD232 pKa = 3.61TPEE235 pKa = 5.51DD236 pKa = 3.9PTHH239 pKa = 7.31DD240 pKa = 4.39PDD242 pKa = 5.96DD243 pKa = 4.23PTDD246 pKa = 4.15EE247 pKa = 5.35IEE249 pKa = 5.27DD250 pKa = 3.98PTVLPDD256 pKa = 4.76DD257 pKa = 3.92STNTVNPGVVDD268 pKa = 4.59DD269 pKa = 5.26KK270 pKa = 11.58PDD272 pKa = 3.54VEE274 pKa = 6.19DD275 pKa = 5.25PDD277 pKa = 4.19TDD279 pKa = 3.44EE280 pKa = 4.48STDD283 pKa = 3.65TAVLSAIKK291 pKa = 10.32GLNVDD296 pKa = 3.91VNKK299 pKa = 10.75GIHH302 pKa = 6.7DD303 pKa = 4.72LNVDD307 pKa = 3.85INQSHH312 pKa = 7.08ADD314 pKa = 3.21ITNAVIDD321 pKa = 4.2VKK323 pKa = 11.2GSLVDD328 pKa = 3.34NTQAIQEE335 pKa = 4.02QQINDD340 pKa = 3.12NKK342 pKa = 10.54IYY344 pKa = 10.9NNTKK348 pKa = 10.74ALIQQANGDD357 pKa = 3.16ITTAVNNNTNATIGIRR373 pKa = 11.84DD374 pKa = 4.0DD375 pKa = 4.27LKK377 pKa = 11.52GLGDD381 pKa = 3.9SVGEE385 pKa = 4.31LDD387 pKa = 4.0SSLNAIEE394 pKa = 4.64GLLTGSEE401 pKa = 4.55FGTPTGTAITGEE413 pKa = 4.12IFTAEE418 pKa = 4.26DD419 pKa = 3.57FANLQTTIDD428 pKa = 3.92EE429 pKa = 4.45KK430 pKa = 11.47AEE432 pKa = 4.29SIQGYY437 pKa = 9.97VDD439 pKa = 4.67DD440 pKa = 4.85IKK442 pKa = 11.59GLITIGTNFNNGTLSDD458 pKa = 3.38KK459 pKa = 11.13SFNIKK464 pKa = 10.03GATVEE469 pKa = 4.33SGLQRR474 pKa = 11.84FDD476 pKa = 3.62AVSGYY481 pKa = 9.57VRR483 pKa = 11.84PVVLFICALIALWVLFGNRR502 pKa = 11.84SKK504 pKa = 11.51
MM1 pKa = 7.28NKK3 pKa = 10.06SLFLLLFSCLFLSLNASAAQPTYY26 pKa = 10.54KK27 pKa = 10.5VSDD30 pKa = 3.43VSAYY34 pKa = 9.5PDD36 pKa = 3.59CKK38 pKa = 10.69LLLGLRR44 pKa = 11.84VNPASYY50 pKa = 10.56VSCYY54 pKa = 8.61EE55 pKa = 4.05NKK57 pKa = 9.73FVNYY61 pKa = 9.99KK62 pKa = 10.62DD63 pKa = 4.74FSTKK67 pKa = 9.71SCYY70 pKa = 9.75LRR72 pKa = 11.84HH73 pKa = 5.84GKK75 pKa = 9.97YY76 pKa = 9.97VVDD79 pKa = 4.27IMCHH83 pKa = 4.35TTSPSWPLYY92 pKa = 9.22RR93 pKa = 11.84AAGFLQNSAQCPPDD107 pKa = 3.53YY108 pKa = 11.08EE109 pKa = 4.4KK110 pKa = 11.45VEE112 pKa = 4.15DD113 pKa = 4.48EE114 pKa = 4.63YY115 pKa = 11.89VVSCEE120 pKa = 4.44PIVPACEE127 pKa = 3.92YY128 pKa = 11.15GEE130 pKa = 4.53NPDD133 pKa = 4.02GSCMDD138 pKa = 3.64ACQFKK143 pKa = 10.73QSIGDD148 pKa = 3.7TVKK151 pKa = 10.86LYY153 pKa = 8.08WHH155 pKa = 6.48PAIYY159 pKa = 10.72GEE161 pKa = 4.73LVTGACYY168 pKa = 10.65GDD170 pKa = 4.13YY171 pKa = 10.89GATRR175 pKa = 11.84CEE177 pKa = 4.02VTKK180 pKa = 11.03NEE182 pKa = 4.01STIICTGVPDD192 pKa = 4.14GQYY195 pKa = 10.09TPDD198 pKa = 3.67SQCSLRR204 pKa = 11.84FAYY207 pKa = 9.17TGRR210 pKa = 11.84QCDD213 pKa = 3.3GGTLFWGVNGPDD225 pKa = 3.77EE226 pKa = 5.2PIIPPDD232 pKa = 3.61TPEE235 pKa = 5.51DD236 pKa = 3.9PTHH239 pKa = 7.31DD240 pKa = 4.39PDD242 pKa = 5.96DD243 pKa = 4.23PTDD246 pKa = 4.15EE247 pKa = 5.35IEE249 pKa = 5.27DD250 pKa = 3.98PTVLPDD256 pKa = 4.76DD257 pKa = 3.92STNTVNPGVVDD268 pKa = 4.59DD269 pKa = 5.26KK270 pKa = 11.58PDD272 pKa = 3.54VEE274 pKa = 6.19DD275 pKa = 5.25PDD277 pKa = 4.19TDD279 pKa = 3.44EE280 pKa = 4.48STDD283 pKa = 3.65TAVLSAIKK291 pKa = 10.32GLNVDD296 pKa = 3.91VNKK299 pKa = 10.75GIHH302 pKa = 6.7DD303 pKa = 4.72LNVDD307 pKa = 3.85INQSHH312 pKa = 7.08ADD314 pKa = 3.21ITNAVIDD321 pKa = 4.2VKK323 pKa = 11.2GSLVDD328 pKa = 3.34NTQAIQEE335 pKa = 4.02QQINDD340 pKa = 3.12NKK342 pKa = 10.54IYY344 pKa = 10.9NNTKK348 pKa = 10.74ALIQQANGDD357 pKa = 3.16ITTAVNNNTNATIGIRR373 pKa = 11.84DD374 pKa = 4.0DD375 pKa = 4.27LKK377 pKa = 11.52GLGDD381 pKa = 3.9SVGEE385 pKa = 4.31LDD387 pKa = 4.0SSLNAIEE394 pKa = 4.64GLLTGSEE401 pKa = 4.55FGTPTGTAITGEE413 pKa = 4.12IFTAEE418 pKa = 4.26DD419 pKa = 3.57FANLQTTIDD428 pKa = 3.92EE429 pKa = 4.45KK430 pKa = 11.47AEE432 pKa = 4.29SIQGYY437 pKa = 9.97VDD439 pKa = 4.67DD440 pKa = 4.85IKK442 pKa = 11.59GLITIGTNFNNGTLSDD458 pKa = 3.38KK459 pKa = 11.13SFNIKK464 pKa = 10.03GATVEE469 pKa = 4.33SGLQRR474 pKa = 11.84FDD476 pKa = 3.62AVSGYY481 pKa = 9.57VRR483 pKa = 11.84PVVLFICALIALWVLFGNRR502 pKa = 11.84SKK504 pKa = 11.51
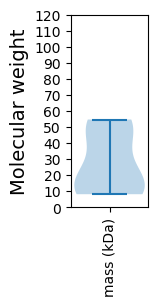
Molecular weight: 54.64 kDa
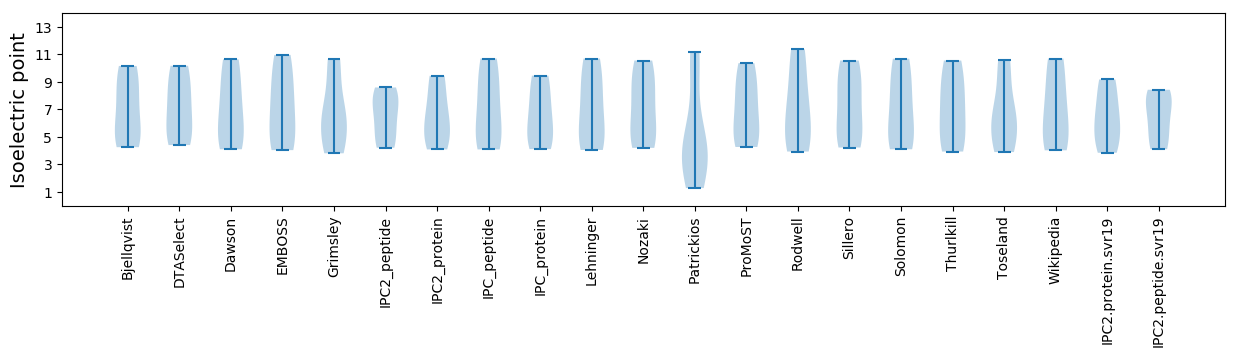
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9MCC2|Q9MCC2_9VIRU Vpf215 OS=Vibrio phage VfO4K68 OX=127508 GN=vpf215 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.36GLDD5 pKa = 3.31MKK7 pKa = 11.07YY8 pKa = 9.36VFRR11 pKa = 11.84GKK13 pKa = 10.48SYY15 pKa = 7.69TTLTSCYY22 pKa = 9.68RR23 pKa = 11.84DD24 pKa = 3.22NTDD27 pKa = 2.87QVTVGIGTVRR37 pKa = 11.84TRR39 pKa = 11.84LKK41 pKa = 11.02NGWSLNKK48 pKa = 10.24ALLHH52 pKa = 6.2PKK54 pKa = 10.02QKK56 pKa = 9.7TIKK59 pKa = 8.75TKK61 pKa = 10.61LGSHH65 pKa = 4.77TVEE68 pKa = 3.85GKK70 pKa = 10.6VYY72 pKa = 10.6EE73 pKa = 4.2NLPSIAEE80 pKa = 4.22EE81 pKa = 4.11YY82 pKa = 11.43GMTLNTIYY90 pKa = 10.55KK91 pKa = 9.43RR92 pKa = 11.84YY93 pKa = 10.22SRR95 pKa = 11.84GCRR98 pKa = 11.84GDD100 pKa = 4.28DD101 pKa = 3.72LVPLKK106 pKa = 10.5KK107 pKa = 9.97RR108 pKa = 11.84KK109 pKa = 10.15SYY111 pKa = 10.07VAPDD115 pKa = 3.53DD116 pKa = 3.58EE117 pKa = 6.1ANFRR121 pKa = 11.84FYY123 pKa = 11.6ANGVGYY129 pKa = 10.55KK130 pKa = 9.83SAADD134 pKa = 3.47ACRR137 pKa = 11.84KK138 pKa = 10.2LNVKK142 pKa = 10.03YY143 pKa = 8.94GTYY146 pKa = 9.93RR147 pKa = 11.84LHH149 pKa = 6.38MSNGFTVEE157 pKa = 3.77QALGIEE163 pKa = 4.44KK164 pKa = 10.43VQDD167 pKa = 3.24GRR169 pKa = 11.84KK170 pKa = 9.56VRR172 pKa = 11.84GTKK175 pKa = 10.3FNVDD179 pKa = 2.89GKK181 pKa = 10.77EE182 pKa = 3.81YY183 pKa = 10.13TINEE187 pKa = 3.99LSILHH192 pKa = 5.63KK193 pKa = 10.32VVRR196 pKa = 11.84NWSRR200 pKa = 11.84RR201 pKa = 11.84FHH203 pKa = 6.02EE204 pKa = 4.54RR205 pKa = 11.84SSNHH209 pKa = 6.24LLRR212 pKa = 11.84LATT215 pKa = 4.07
MM1 pKa = 7.59KK2 pKa = 10.36GLDD5 pKa = 3.31MKK7 pKa = 11.07YY8 pKa = 9.36VFRR11 pKa = 11.84GKK13 pKa = 10.48SYY15 pKa = 7.69TTLTSCYY22 pKa = 9.68RR23 pKa = 11.84DD24 pKa = 3.22NTDD27 pKa = 2.87QVTVGIGTVRR37 pKa = 11.84TRR39 pKa = 11.84LKK41 pKa = 11.02NGWSLNKK48 pKa = 10.24ALLHH52 pKa = 6.2PKK54 pKa = 10.02QKK56 pKa = 9.7TIKK59 pKa = 8.75TKK61 pKa = 10.61LGSHH65 pKa = 4.77TVEE68 pKa = 3.85GKK70 pKa = 10.6VYY72 pKa = 10.6EE73 pKa = 4.2NLPSIAEE80 pKa = 4.22EE81 pKa = 4.11YY82 pKa = 11.43GMTLNTIYY90 pKa = 10.55KK91 pKa = 9.43RR92 pKa = 11.84YY93 pKa = 10.22SRR95 pKa = 11.84GCRR98 pKa = 11.84GDD100 pKa = 4.28DD101 pKa = 3.72LVPLKK106 pKa = 10.5KK107 pKa = 9.97RR108 pKa = 11.84KK109 pKa = 10.15SYY111 pKa = 10.07VAPDD115 pKa = 3.53DD116 pKa = 3.58EE117 pKa = 6.1ANFRR121 pKa = 11.84FYY123 pKa = 11.6ANGVGYY129 pKa = 10.55KK130 pKa = 9.83SAADD134 pKa = 3.47ACRR137 pKa = 11.84KK138 pKa = 10.2LNVKK142 pKa = 10.03YY143 pKa = 8.94GTYY146 pKa = 9.93RR147 pKa = 11.84LHH149 pKa = 6.38MSNGFTVEE157 pKa = 3.77QALGIEE163 pKa = 4.44KK164 pKa = 10.43VQDD167 pKa = 3.24GRR169 pKa = 11.84KK170 pKa = 9.56VRR172 pKa = 11.84GTKK175 pKa = 10.3FNVDD179 pKa = 2.89GKK181 pKa = 10.77EE182 pKa = 3.81YY183 pKa = 10.13TINEE187 pKa = 3.99LSILHH192 pKa = 5.63KK193 pKa = 10.32VVRR196 pKa = 11.84NWSRR200 pKa = 11.84RR201 pKa = 11.84FHH203 pKa = 6.02EE204 pKa = 4.54RR205 pKa = 11.84SSNHH209 pKa = 6.24LLRR212 pKa = 11.84LATT215 pKa = 4.07
Molecular weight: 24.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1972 |
76 |
504 |
246.5 |
27.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.846 ± 0.716 | 2.079 ± 0.442 |
6.389 ± 0.914 | 5.325 ± 0.454 |
4.513 ± 0.583 | 7.606 ± 0.443 |
2.282 ± 0.48 | 5.527 ± 0.608 |
5.477 ± 0.715 | 8.671 ± 0.428 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.724 ± 0.41 | 5.02 ± 0.489 |
4.26 ± 0.632 | 3.347 ± 0.415 |
4.564 ± 0.875 | 6.491 ± 0.426 |
6.643 ± 0.617 | 7.657 ± 0.859 |
1.471 ± 0.281 | 4.108 ± 0.423 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
