
Avon-Heathcote Estuary associated circular virus 27
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
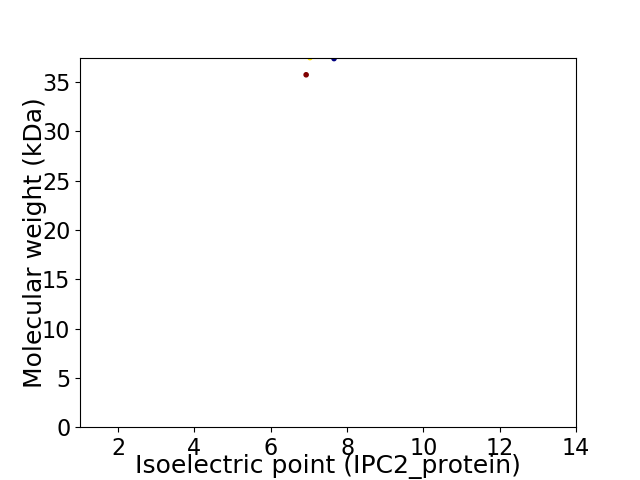
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IBU2|A0A0C5IBU2_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 27 OX=1618251 PE=4 SV=1
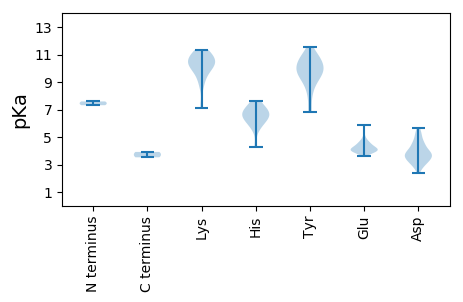
MM1 pKa = 7.59PAKK4 pKa = 10.08QAFNMPFTAFDD15 pKa = 3.63VEE17 pKa = 4.53DD18 pKa = 4.43IEE20 pKa = 4.78WKK22 pKa = 9.06PDD24 pKa = 2.87HH25 pKa = 6.56MKK27 pKa = 10.43YY28 pKa = 10.61LLYY31 pKa = 10.78GRR33 pKa = 11.84EE34 pKa = 4.27TCPSTGRR41 pKa = 11.84EE42 pKa = 3.89HH43 pKa = 6.56LQGYY47 pKa = 6.99VQFIKK52 pKa = 10.59KK53 pKa = 7.28QTIAGAKK60 pKa = 9.9KK61 pKa = 9.86IFDD64 pKa = 3.45NATIHH69 pKa = 6.96FEE71 pKa = 4.09FALGDD76 pKa = 3.53YY77 pKa = 10.46EE78 pKa = 5.86EE79 pKa = 4.24NFKK82 pKa = 11.34YY83 pKa = 9.28CTKK86 pKa = 10.36TRR88 pKa = 11.84VNDD91 pKa = 3.62MVPNVYY97 pKa = 10.06FEE99 pKa = 4.58RR100 pKa = 11.84GEE102 pKa = 3.93PTRR105 pKa = 11.84KK106 pKa = 9.58KK107 pKa = 10.47GQRR110 pKa = 11.84NDD112 pKa = 2.9IQEE115 pKa = 4.04VIRR118 pKa = 11.84RR119 pKa = 11.84IEE121 pKa = 4.02SGNDD125 pKa = 3.0LRR127 pKa = 11.84DD128 pKa = 3.36LMFDD132 pKa = 3.8DD133 pKa = 5.24EE134 pKa = 5.03VNQTVARR141 pKa = 11.84CMPFVRR147 pKa = 11.84QAIADD152 pKa = 4.64FKK154 pKa = 11.09AGKK157 pKa = 9.41GLSAIKK163 pKa = 10.47SKK165 pKa = 11.27LEE167 pKa = 3.92SAQLRR172 pKa = 11.84SWQHH176 pKa = 4.24GVKK179 pKa = 10.29SLVEE183 pKa = 3.9QEE185 pKa = 3.98PEE187 pKa = 3.91GRR189 pKa = 11.84SVYY192 pKa = 9.83WYY194 pKa = 9.46WDD196 pKa = 3.4TKK198 pKa = 11.19GNTGKK203 pKa = 10.7SFMVDD208 pKa = 2.38WLVAFHH214 pKa = 6.65KK215 pKa = 10.78AVVFTHH221 pKa = 6.24GKK223 pKa = 7.14MHH225 pKa = 7.6DD226 pKa = 3.17IAHH229 pKa = 7.38AYY231 pKa = 8.11NHH233 pKa = 6.14EE234 pKa = 4.18KK235 pKa = 10.4VVCFDD240 pKa = 3.99LSRR243 pKa = 11.84TQEE246 pKa = 3.91EE247 pKa = 4.53KK248 pKa = 11.09LDD250 pKa = 3.8AVYY253 pKa = 9.14MALEE257 pKa = 4.23CFKK260 pKa = 10.65NGRR263 pKa = 11.84MFSGKK268 pKa = 10.08YY269 pKa = 9.81EE270 pKa = 4.09STTKK274 pKa = 10.21IFEE277 pKa = 4.32VPHH280 pKa = 5.73VFVFANFAPDD290 pKa = 3.36KK291 pKa = 10.22TKK293 pKa = 10.85LSEE296 pKa = 4.39DD297 pKa = 2.93RR298 pKa = 11.84WKK300 pKa = 9.29VTEE303 pKa = 4.75IISFF307 pKa = 3.92
MM1 pKa = 7.59PAKK4 pKa = 10.08QAFNMPFTAFDD15 pKa = 3.63VEE17 pKa = 4.53DD18 pKa = 4.43IEE20 pKa = 4.78WKK22 pKa = 9.06PDD24 pKa = 2.87HH25 pKa = 6.56MKK27 pKa = 10.43YY28 pKa = 10.61LLYY31 pKa = 10.78GRR33 pKa = 11.84EE34 pKa = 4.27TCPSTGRR41 pKa = 11.84EE42 pKa = 3.89HH43 pKa = 6.56LQGYY47 pKa = 6.99VQFIKK52 pKa = 10.59KK53 pKa = 7.28QTIAGAKK60 pKa = 9.9KK61 pKa = 9.86IFDD64 pKa = 3.45NATIHH69 pKa = 6.96FEE71 pKa = 4.09FALGDD76 pKa = 3.53YY77 pKa = 10.46EE78 pKa = 5.86EE79 pKa = 4.24NFKK82 pKa = 11.34YY83 pKa = 9.28CTKK86 pKa = 10.36TRR88 pKa = 11.84VNDD91 pKa = 3.62MVPNVYY97 pKa = 10.06FEE99 pKa = 4.58RR100 pKa = 11.84GEE102 pKa = 3.93PTRR105 pKa = 11.84KK106 pKa = 9.58KK107 pKa = 10.47GQRR110 pKa = 11.84NDD112 pKa = 2.9IQEE115 pKa = 4.04VIRR118 pKa = 11.84RR119 pKa = 11.84IEE121 pKa = 4.02SGNDD125 pKa = 3.0LRR127 pKa = 11.84DD128 pKa = 3.36LMFDD132 pKa = 3.8DD133 pKa = 5.24EE134 pKa = 5.03VNQTVARR141 pKa = 11.84CMPFVRR147 pKa = 11.84QAIADD152 pKa = 4.64FKK154 pKa = 11.09AGKK157 pKa = 9.41GLSAIKK163 pKa = 10.47SKK165 pKa = 11.27LEE167 pKa = 3.92SAQLRR172 pKa = 11.84SWQHH176 pKa = 4.24GVKK179 pKa = 10.29SLVEE183 pKa = 3.9QEE185 pKa = 3.98PEE187 pKa = 3.91GRR189 pKa = 11.84SVYY192 pKa = 9.83WYY194 pKa = 9.46WDD196 pKa = 3.4TKK198 pKa = 11.19GNTGKK203 pKa = 10.7SFMVDD208 pKa = 2.38WLVAFHH214 pKa = 6.65KK215 pKa = 10.78AVVFTHH221 pKa = 6.24GKK223 pKa = 7.14MHH225 pKa = 7.6DD226 pKa = 3.17IAHH229 pKa = 7.38AYY231 pKa = 8.11NHH233 pKa = 6.14EE234 pKa = 4.18KK235 pKa = 10.4VVCFDD240 pKa = 3.99LSRR243 pKa = 11.84TQEE246 pKa = 3.91EE247 pKa = 4.53KK248 pKa = 11.09LDD250 pKa = 3.8AVYY253 pKa = 9.14MALEE257 pKa = 4.23CFKK260 pKa = 10.65NGRR263 pKa = 11.84MFSGKK268 pKa = 10.08YY269 pKa = 9.81EE270 pKa = 4.09STTKK274 pKa = 10.21IFEE277 pKa = 4.32VPHH280 pKa = 5.73VFVFANFAPDD290 pKa = 3.36KK291 pKa = 10.22TKK293 pKa = 10.85LSEE296 pKa = 4.39DD297 pKa = 2.93RR298 pKa = 11.84WKK300 pKa = 9.29VTEE303 pKa = 4.75IISFF307 pKa = 3.92
Molecular weight: 35.73 kDa
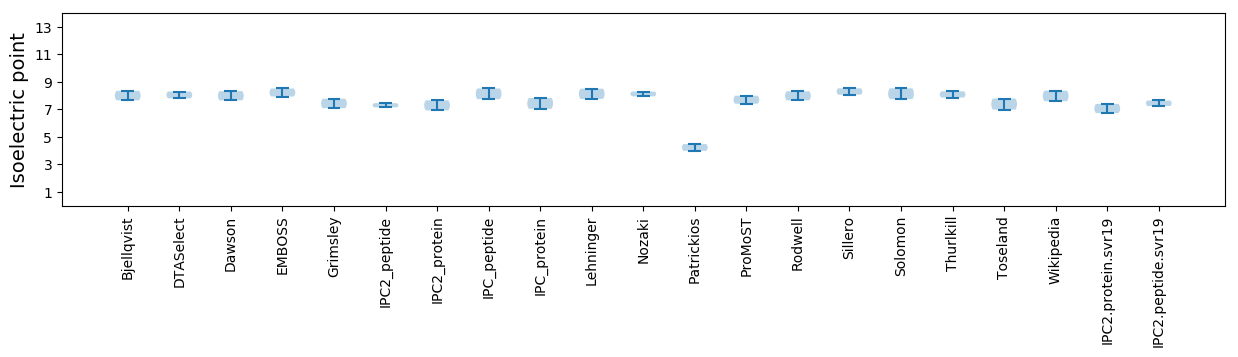
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IBU2|A0A0C5IBU2_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 27 OX=1618251 PE=4 SV=1
MM1 pKa = 7.31KK2 pKa = 10.18RR3 pKa = 11.84ARR5 pKa = 11.84DD6 pKa = 3.77YY7 pKa = 10.96QRR9 pKa = 11.84PRR11 pKa = 11.84YY12 pKa = 8.85SKK14 pKa = 10.01RR15 pKa = 11.84SRR17 pKa = 11.84RR18 pKa = 11.84PTPQRR23 pKa = 11.84SMRR26 pKa = 11.84SVIGRR31 pKa = 11.84LAEE34 pKa = 4.13SKK36 pKa = 10.66RR37 pKa = 11.84EE38 pKa = 3.91NFHH41 pKa = 7.13VSSQTLNHH49 pKa = 7.03DD50 pKa = 3.9AGSGQVVGGFARR62 pKa = 11.84PNNRR66 pKa = 11.84SGFNWMGLNQGDD78 pKa = 4.36SKK80 pKa = 11.07HH81 pKa = 6.86DD82 pKa = 3.81RR83 pKa = 11.84DD84 pKa = 3.68GDD86 pKa = 4.54SIFSQYY92 pKa = 9.93IDD94 pKa = 3.86YY95 pKa = 10.83KK96 pKa = 10.87LQLTGYY102 pKa = 9.69SDD104 pKa = 4.46YY105 pKa = 10.77PSQSCRR111 pKa = 11.84VIIFTPKK118 pKa = 10.45ALRR121 pKa = 11.84TNTLIASSNVASLFVTDD138 pKa = 5.28GSDD141 pKa = 3.29SGSGNLLIQPVDD153 pKa = 3.42TFNYY157 pKa = 9.55NVLRR161 pKa = 11.84DD162 pKa = 4.12FVVHH166 pKa = 6.35PLKK169 pKa = 10.88NQNQNSSAAQSTGLLYY185 pKa = 10.67DD186 pKa = 3.54VLHH189 pKa = 6.75PSISSGPFTSVAGTYY204 pKa = 9.86RR205 pKa = 11.84EE206 pKa = 4.48LFNKK210 pKa = 8.76YY211 pKa = 10.06HH212 pKa = 6.31SIIYY216 pKa = 10.22NEE218 pKa = 4.09LPALEE223 pKa = 4.54SAAPTLAPYY232 pKa = 9.93IAAKK236 pKa = 9.93IAEE239 pKa = 4.18SQYY242 pKa = 11.06YY243 pKa = 8.84INEE246 pKa = 4.12YY247 pKa = 10.15VDD249 pKa = 5.63DD250 pKa = 3.86VTEE253 pKa = 3.98YY254 pKa = 11.58SNIPFDD260 pKa = 4.29PVDD263 pKa = 4.87DD264 pKa = 5.18AGNDD268 pKa = 3.38CNYY271 pKa = 9.54PIVSGRR277 pKa = 11.84IKK279 pKa = 9.59TNRR282 pKa = 11.84RR283 pKa = 11.84VQYY286 pKa = 8.12EE287 pKa = 4.17TDD289 pKa = 3.07SVRR292 pKa = 11.84PLKK295 pKa = 8.62TTDD298 pKa = 4.6FIQIAIIPYY307 pKa = 9.14ADD309 pKa = 3.84PSAVTTTTIMRR320 pKa = 11.84YY321 pKa = 6.79NQEE324 pKa = 3.61VTHH327 pKa = 5.66YY328 pKa = 10.43FKK330 pKa = 11.21DD331 pKa = 3.91FF332 pKa = 3.52
MM1 pKa = 7.31KK2 pKa = 10.18RR3 pKa = 11.84ARR5 pKa = 11.84DD6 pKa = 3.77YY7 pKa = 10.96QRR9 pKa = 11.84PRR11 pKa = 11.84YY12 pKa = 8.85SKK14 pKa = 10.01RR15 pKa = 11.84SRR17 pKa = 11.84RR18 pKa = 11.84PTPQRR23 pKa = 11.84SMRR26 pKa = 11.84SVIGRR31 pKa = 11.84LAEE34 pKa = 4.13SKK36 pKa = 10.66RR37 pKa = 11.84EE38 pKa = 3.91NFHH41 pKa = 7.13VSSQTLNHH49 pKa = 7.03DD50 pKa = 3.9AGSGQVVGGFARR62 pKa = 11.84PNNRR66 pKa = 11.84SGFNWMGLNQGDD78 pKa = 4.36SKK80 pKa = 11.07HH81 pKa = 6.86DD82 pKa = 3.81RR83 pKa = 11.84DD84 pKa = 3.68GDD86 pKa = 4.54SIFSQYY92 pKa = 9.93IDD94 pKa = 3.86YY95 pKa = 10.83KK96 pKa = 10.87LQLTGYY102 pKa = 9.69SDD104 pKa = 4.46YY105 pKa = 10.77PSQSCRR111 pKa = 11.84VIIFTPKK118 pKa = 10.45ALRR121 pKa = 11.84TNTLIASSNVASLFVTDD138 pKa = 5.28GSDD141 pKa = 3.29SGSGNLLIQPVDD153 pKa = 3.42TFNYY157 pKa = 9.55NVLRR161 pKa = 11.84DD162 pKa = 4.12FVVHH166 pKa = 6.35PLKK169 pKa = 10.88NQNQNSSAAQSTGLLYY185 pKa = 10.67DD186 pKa = 3.54VLHH189 pKa = 6.75PSISSGPFTSVAGTYY204 pKa = 9.86RR205 pKa = 11.84EE206 pKa = 4.48LFNKK210 pKa = 8.76YY211 pKa = 10.06HH212 pKa = 6.31SIIYY216 pKa = 10.22NEE218 pKa = 4.09LPALEE223 pKa = 4.54SAAPTLAPYY232 pKa = 9.93IAAKK236 pKa = 9.93IAEE239 pKa = 4.18SQYY242 pKa = 11.06YY243 pKa = 8.84INEE246 pKa = 4.12YY247 pKa = 10.15VDD249 pKa = 5.63DD250 pKa = 3.86VTEE253 pKa = 3.98YY254 pKa = 11.58SNIPFDD260 pKa = 4.29PVDD263 pKa = 4.87DD264 pKa = 5.18AGNDD268 pKa = 3.38CNYY271 pKa = 9.54PIVSGRR277 pKa = 11.84IKK279 pKa = 9.59TNRR282 pKa = 11.84RR283 pKa = 11.84VQYY286 pKa = 8.12EE287 pKa = 4.17TDD289 pKa = 3.07SVRR292 pKa = 11.84PLKK295 pKa = 8.62TTDD298 pKa = 4.6FIQIAIIPYY307 pKa = 9.14ADD309 pKa = 3.84PSAVTTTTIMRR320 pKa = 11.84YY321 pKa = 6.79NQEE324 pKa = 3.61VTHH327 pKa = 5.66YY328 pKa = 10.43FKK330 pKa = 11.21DD331 pKa = 3.91FF332 pKa = 3.52
Molecular weight: 37.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
639 |
307 |
332 |
319.5 |
36.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.573 ± 0.185 | 1.095 ± 0.368 |
6.573 ± 0.265 | 5.321 ± 1.723 |
5.79 ± 1.174 | 5.477 ± 0.042 |
2.66 ± 0.412 | 5.477 ± 0.633 |
6.26 ± 1.974 | 5.477 ± 0.408 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.191 ± 0.736 | 5.321 ± 0.974 |
4.538 ± 0.884 | 4.382 ± 0.326 |
6.103 ± 0.615 | 7.668 ± 2.145 |
6.103 ± 0.39 | 6.886 ± 0.418 |
1.095 ± 0.593 | 5.008 ± 0.983 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
