
Human papillomavirus 141
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 11
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
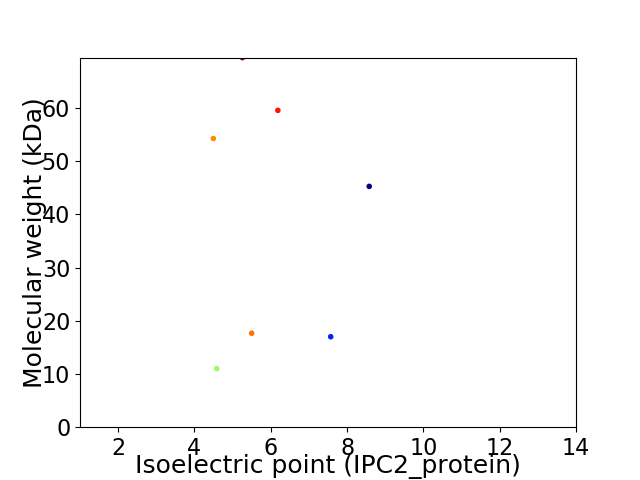
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
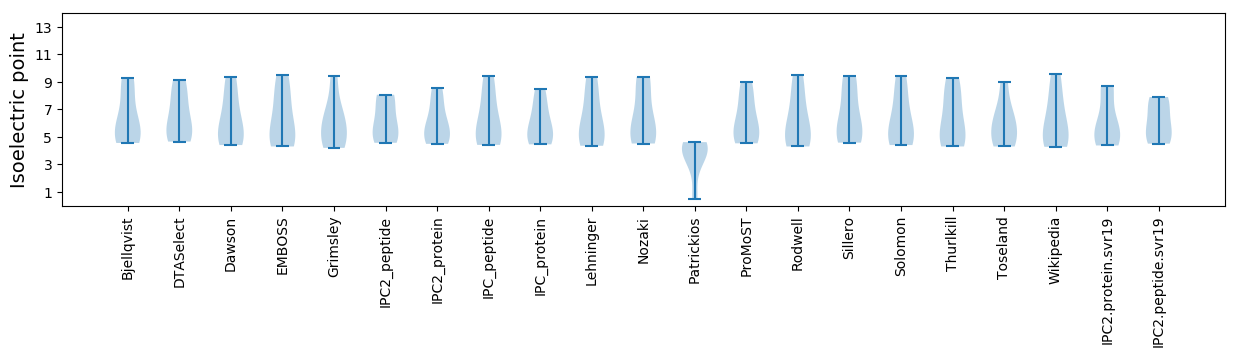
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3P6N3|I3P6N3_9PAPI Replication protein E1 OS=Human papillomavirus 141 OX=1070414 GN=E1 PE=3 SV=1
MM1 pKa = 7.1MGEE4 pKa = 4.44KK5 pKa = 9.07PTMRR9 pKa = 11.84DD10 pKa = 2.43IDD12 pKa = 4.09LEE14 pKa = 4.29EE15 pKa = 4.08TLEE18 pKa = 4.0NLVMPSSLLCEE29 pKa = 4.44EE30 pKa = 4.74SLSPDD35 pKa = 3.34DD36 pKa = 4.0TPEE39 pKa = 4.08EE40 pKa = 4.39EE41 pKa = 4.46SLSPYY46 pKa = 9.32WVDD49 pKa = 4.87SRR51 pKa = 11.84CDD53 pKa = 3.19SCKK56 pKa = 10.72NPVRR60 pKa = 11.84LCVVATSGAIHH71 pKa = 7.14LLEE74 pKa = 3.76QLLFNDD80 pKa = 4.52LCIVCPVCSRR90 pKa = 11.84TLVRR94 pKa = 11.84HH95 pKa = 5.63GRR97 pKa = 11.84HH98 pKa = 5.27
MM1 pKa = 7.1MGEE4 pKa = 4.44KK5 pKa = 9.07PTMRR9 pKa = 11.84DD10 pKa = 2.43IDD12 pKa = 4.09LEE14 pKa = 4.29EE15 pKa = 4.08TLEE18 pKa = 4.0NLVMPSSLLCEE29 pKa = 4.44EE30 pKa = 4.74SLSPDD35 pKa = 3.34DD36 pKa = 4.0TPEE39 pKa = 4.08EE40 pKa = 4.39EE41 pKa = 4.46SLSPYY46 pKa = 9.32WVDD49 pKa = 4.87SRR51 pKa = 11.84CDD53 pKa = 3.19SCKK56 pKa = 10.72NPVRR60 pKa = 11.84LCVVATSGAIHH71 pKa = 7.14LLEE74 pKa = 3.76QLLFNDD80 pKa = 4.52LCIVCPVCSRR90 pKa = 11.84TLVRR94 pKa = 11.84HH95 pKa = 5.63GRR97 pKa = 11.84HH98 pKa = 5.27
Molecular weight: 11.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3P6N5|I3P6N5_9PAPI Uncharacterized protein OS=Human papillomavirus 141 OX=1070414 GN=E4 PE=4 SV=1
MM1 pKa = 6.88NQEE4 pKa = 4.39DD5 pKa = 3.85LRR7 pKa = 11.84KK8 pKa = 9.71RR9 pKa = 11.84LDD11 pKa = 3.69VIQTQLMTLYY21 pKa = 10.41EE22 pKa = 4.7SNPKK26 pKa = 10.05DD27 pKa = 3.56LQSQITHH34 pKa = 5.8YY35 pKa = 10.96KK36 pKa = 10.24LLRR39 pKa = 11.84KK40 pKa = 9.31EE41 pKa = 4.3CAIQYY46 pKa = 6.75YY47 pKa = 9.77ARR49 pKa = 11.84KK50 pKa = 9.56EE51 pKa = 4.33GYY53 pKa = 9.79QNLGLQHH60 pKa = 7.13LPTTRR65 pKa = 11.84ISEE68 pKa = 4.44HH69 pKa = 6.16NSKK72 pKa = 10.24QAIKK76 pKa = 7.91MTLMLEE82 pKa = 4.27SLAKK86 pKa = 9.85SQYY89 pKa = 9.15ATEE92 pKa = 4.03EE93 pKa = 4.09WTLHH97 pKa = 5.67DD98 pKa = 3.59TSADD102 pKa = 3.54RR103 pKa = 11.84FLSPPRR109 pKa = 11.84NCFKK113 pKa = 10.94KK114 pKa = 10.35NSFEE118 pKa = 4.29VEE120 pKa = 3.72VWFDD124 pKa = 3.98NNPKK128 pKa = 10.26NAFPYY133 pKa = 10.07ICWEE137 pKa = 3.99WIYY140 pKa = 11.09YY141 pKa = 9.18QDD143 pKa = 5.77EE144 pKa = 4.34SDD146 pKa = 3.43TWHH149 pKa = 6.76KK150 pKa = 11.21VPGKK154 pKa = 10.14VDD156 pKa = 3.61YY157 pKa = 11.26NGLYY161 pKa = 9.88YY162 pKa = 10.62VEE164 pKa = 4.54IDD166 pKa = 3.22GTAVYY171 pKa = 10.08FLLFHH176 pKa = 7.19KK177 pKa = 10.78DD178 pKa = 2.6AGRR181 pKa = 11.84YY182 pKa = 8.3GNSGEE187 pKa = 3.85WTVNYY192 pKa = 10.03KK193 pKa = 10.32NEE195 pKa = 4.26QILPPSVGSSTRR207 pKa = 11.84RR208 pKa = 11.84SVSEE212 pKa = 4.14SQDD215 pKa = 3.15PTINTPSNAEE225 pKa = 3.87TEE227 pKa = 4.43TQNRR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84EE234 pKa = 4.24VQQEE238 pKa = 4.35TPGPSSTTRR247 pKa = 11.84SPKK250 pKa = 8.97RR251 pKa = 11.84GQRR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84GGEE260 pKa = 3.91GEE262 pKa = 3.98QTSGKK267 pKa = 9.37RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84SEE273 pKa = 3.64GRR275 pKa = 11.84GGSDD279 pKa = 3.15SITPEE284 pKa = 3.99EE285 pKa = 4.12VGKK288 pKa = 8.31SHH290 pKa = 7.38RR291 pKa = 11.84SVPATGLSRR300 pKa = 11.84VDD302 pKa = 3.34RR303 pKa = 11.84LKK305 pKa = 11.26AEE307 pKa = 4.54AKK309 pKa = 9.81DD310 pKa = 3.57PPIIIVKK317 pKa = 10.3GCANKK322 pKa = 10.1LKK324 pKa = 10.33CWRR327 pKa = 11.84FRR329 pKa = 11.84CNQKK333 pKa = 10.12CPRR336 pKa = 11.84PYY338 pKa = 11.0NYY340 pKa = 7.16MTSVFKK346 pKa = 10.61WITNDD351 pKa = 2.89VKK353 pKa = 11.06LADD356 pKa = 3.84SRR358 pKa = 11.84VLVAFADD365 pKa = 3.49NTQRR369 pKa = 11.84SRR371 pKa = 11.84FIANVSFPKK380 pKa = 9.79DD381 pKa = 3.07TSYY384 pKa = 11.57CFGSLEE390 pKa = 4.18CLL392 pKa = 3.55
MM1 pKa = 6.88NQEE4 pKa = 4.39DD5 pKa = 3.85LRR7 pKa = 11.84KK8 pKa = 9.71RR9 pKa = 11.84LDD11 pKa = 3.69VIQTQLMTLYY21 pKa = 10.41EE22 pKa = 4.7SNPKK26 pKa = 10.05DD27 pKa = 3.56LQSQITHH34 pKa = 5.8YY35 pKa = 10.96KK36 pKa = 10.24LLRR39 pKa = 11.84KK40 pKa = 9.31EE41 pKa = 4.3CAIQYY46 pKa = 6.75YY47 pKa = 9.77ARR49 pKa = 11.84KK50 pKa = 9.56EE51 pKa = 4.33GYY53 pKa = 9.79QNLGLQHH60 pKa = 7.13LPTTRR65 pKa = 11.84ISEE68 pKa = 4.44HH69 pKa = 6.16NSKK72 pKa = 10.24QAIKK76 pKa = 7.91MTLMLEE82 pKa = 4.27SLAKK86 pKa = 9.85SQYY89 pKa = 9.15ATEE92 pKa = 4.03EE93 pKa = 4.09WTLHH97 pKa = 5.67DD98 pKa = 3.59TSADD102 pKa = 3.54RR103 pKa = 11.84FLSPPRR109 pKa = 11.84NCFKK113 pKa = 10.94KK114 pKa = 10.35NSFEE118 pKa = 4.29VEE120 pKa = 3.72VWFDD124 pKa = 3.98NNPKK128 pKa = 10.26NAFPYY133 pKa = 10.07ICWEE137 pKa = 3.99WIYY140 pKa = 11.09YY141 pKa = 9.18QDD143 pKa = 5.77EE144 pKa = 4.34SDD146 pKa = 3.43TWHH149 pKa = 6.76KK150 pKa = 11.21VPGKK154 pKa = 10.14VDD156 pKa = 3.61YY157 pKa = 11.26NGLYY161 pKa = 9.88YY162 pKa = 10.62VEE164 pKa = 4.54IDD166 pKa = 3.22GTAVYY171 pKa = 10.08FLLFHH176 pKa = 7.19KK177 pKa = 10.78DD178 pKa = 2.6AGRR181 pKa = 11.84YY182 pKa = 8.3GNSGEE187 pKa = 3.85WTVNYY192 pKa = 10.03KK193 pKa = 10.32NEE195 pKa = 4.26QILPPSVGSSTRR207 pKa = 11.84RR208 pKa = 11.84SVSEE212 pKa = 4.14SQDD215 pKa = 3.15PTINTPSNAEE225 pKa = 3.87TEE227 pKa = 4.43TQNRR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84EE234 pKa = 4.24VQQEE238 pKa = 4.35TPGPSSTTRR247 pKa = 11.84SPKK250 pKa = 8.97RR251 pKa = 11.84GQRR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84GGEE260 pKa = 3.91GEE262 pKa = 3.98QTSGKK267 pKa = 9.37RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84SEE273 pKa = 3.64GRR275 pKa = 11.84GGSDD279 pKa = 3.15SITPEE284 pKa = 3.99EE285 pKa = 4.12VGKK288 pKa = 8.31SHH290 pKa = 7.38RR291 pKa = 11.84SVPATGLSRR300 pKa = 11.84VDD302 pKa = 3.34RR303 pKa = 11.84LKK305 pKa = 11.26AEE307 pKa = 4.54AKK309 pKa = 9.81DD310 pKa = 3.57PPIIIVKK317 pKa = 10.3GCANKK322 pKa = 10.1LKK324 pKa = 10.33CWRR327 pKa = 11.84FRR329 pKa = 11.84CNQKK333 pKa = 10.12CPRR336 pKa = 11.84PYY338 pKa = 11.0NYY340 pKa = 7.16MTSVFKK346 pKa = 10.61WITNDD351 pKa = 2.89VKK353 pKa = 11.06LADD356 pKa = 3.84SRR358 pKa = 11.84VLVAFADD365 pKa = 3.49NTQRR369 pKa = 11.84SRR371 pKa = 11.84FIANVSFPKK380 pKa = 9.79DD381 pKa = 3.07TSYY384 pKa = 11.57CFGSLEE390 pKa = 4.18CLL392 pKa = 3.55
Molecular weight: 45.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2408 |
98 |
604 |
344.0 |
39.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.191 ± 0.398 | 2.575 ± 0.903 |
6.894 ± 0.478 | 6.063 ± 0.51 |
4.693 ± 0.448 | 4.776 ± 0.847 |
1.91 ± 0.145 | 5.648 ± 0.564 |
5.731 ± 0.836 | 9.302 ± 1.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.703 ± 0.348 | 4.817 ± 0.279 |
6.271 ± 0.994 | 4.734 ± 0.465 |
5.689 ± 0.752 | 7.434 ± 0.629 |
6.312 ± 0.743 | 5.399 ± 0.35 |
1.287 ± 0.232 | 3.571 ± 0.567 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
