
Hubei odonate virus 11
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Jingchuvirales; Chuviridae; Odonatavirus; Odonatavirus fabricii
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
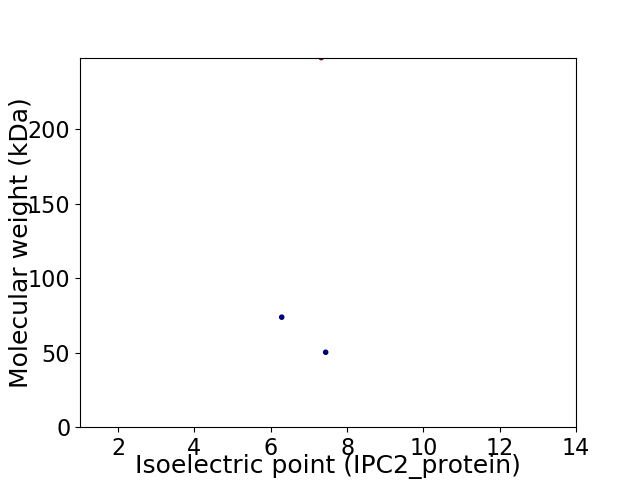
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
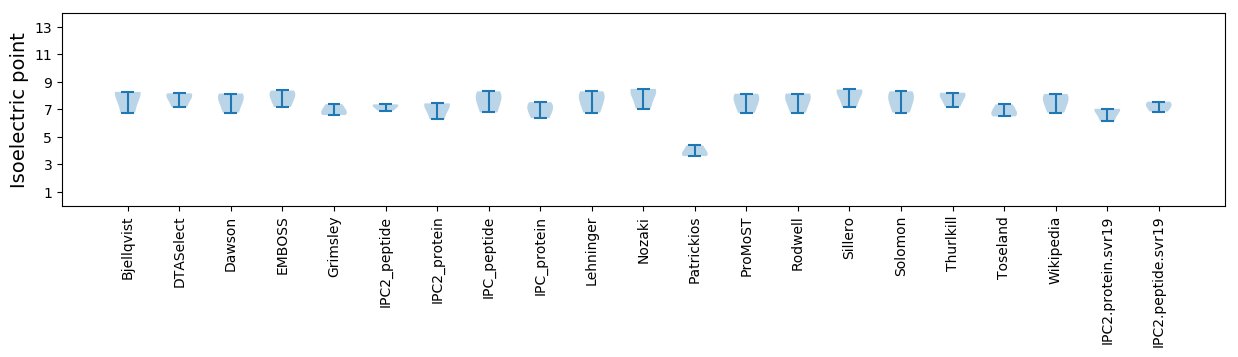
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMU3|A0A1L3KMU3_9VIRU Uncharacterized protein OS=Hubei odonate virus 11 OX=1922992 PE=4 SV=1
MM1 pKa = 7.29YY2 pKa = 9.65IKK4 pKa = 10.58KK5 pKa = 9.84IFWFISLTLCMLEE18 pKa = 3.65ISGLIGFDD26 pKa = 3.8CKK28 pKa = 10.91HH29 pKa = 6.67KK30 pKa = 10.56DD31 pKa = 3.43VEE33 pKa = 4.39YY34 pKa = 10.78SAVSVKK40 pKa = 9.8QVAEE44 pKa = 3.98CDD46 pKa = 3.52EE47 pKa = 4.55VKK49 pKa = 10.54PAVHH53 pKa = 5.57QSKK56 pKa = 10.21MNIQLLQKK64 pKa = 10.21RR65 pKa = 11.84RR66 pKa = 11.84LSKK69 pKa = 10.66VPYY72 pKa = 9.13ISCYY76 pKa = 6.35THH78 pKa = 6.73ANILITHH85 pKa = 7.1CGMHH89 pKa = 5.47SHH91 pKa = 6.8ASVVAGGLSQRR102 pKa = 11.84IEE104 pKa = 4.06QYY106 pKa = 10.28TSEE109 pKa = 4.08TCRR112 pKa = 11.84RR113 pKa = 11.84AFATKK118 pKa = 9.4TVILFGTPIDD128 pKa = 3.53INSFNTTITGSIIPYY143 pKa = 10.01GSVINNDD150 pKa = 3.23ASCIGTSFAYY160 pKa = 10.47NGNHH164 pKa = 4.79YY165 pKa = 9.41RR166 pKa = 11.84ASVMQVSYY174 pKa = 10.77EE175 pKa = 4.19IILKK179 pKa = 8.85TGYY182 pKa = 8.36GTYY185 pKa = 10.47DD186 pKa = 3.28SEE188 pKa = 4.38TGIFNTGLGSYY199 pKa = 10.52GDD201 pKa = 3.84YY202 pKa = 11.03KK203 pKa = 11.06EE204 pKa = 5.59GVLLDD209 pKa = 3.87RR210 pKa = 11.84NFGYY214 pKa = 10.15VFWKK218 pKa = 9.8TSDD221 pKa = 4.18NEE223 pKa = 4.39DD224 pKa = 3.81CSSSTSYY231 pKa = 11.04LVLYY235 pKa = 8.17EE236 pKa = 4.6GPAEE240 pKa = 4.09KK241 pKa = 10.4LVSEE245 pKa = 4.47RR246 pKa = 11.84DD247 pKa = 3.3NRR249 pKa = 11.84SIIVVEE255 pKa = 4.14QQSTTFSLQLRR266 pKa = 11.84EE267 pKa = 4.04PTLLCSQHH275 pKa = 6.36AFNTEE280 pKa = 3.35HH281 pKa = 7.08DD282 pKa = 3.72QLKK285 pKa = 10.54IITSNNRR292 pKa = 11.84MFFFKK297 pKa = 10.61RR298 pKa = 11.84IADD301 pKa = 3.73SLDD304 pKa = 3.43INLSTYY310 pKa = 10.55INAKK314 pKa = 8.1FVYY317 pKa = 9.78VEE319 pKa = 3.55RR320 pKa = 11.84HH321 pKa = 4.48LRR323 pKa = 11.84ANIEE327 pKa = 4.05TLYY330 pKa = 10.44EE331 pKa = 3.87DD332 pKa = 3.63VMKK335 pKa = 10.35FRR337 pKa = 11.84CRR339 pKa = 11.84VTRR342 pKa = 11.84QTLTNLLSLASIDD355 pKa = 3.72EE356 pKa = 4.49DD357 pKa = 3.75EE358 pKa = 4.31FAYY361 pKa = 10.29AYY363 pKa = 8.77MGRR366 pKa = 11.84PGYY369 pKa = 9.38TSVKK373 pKa = 9.14RR374 pKa = 11.84GEE376 pKa = 3.79VAYY379 pKa = 10.31LIKK382 pKa = 10.62CVPTYY387 pKa = 9.59ITLQSSTRR395 pKa = 11.84CYY397 pKa = 8.83NDD399 pKa = 3.06VVISHH404 pKa = 5.84NNKK407 pKa = 8.77TKK409 pKa = 10.59FLASKK414 pKa = 9.43TNIIQDD420 pKa = 3.41FSEE423 pKa = 4.29EE424 pKa = 4.43VEE426 pKa = 4.45CSQLTPVLYY435 pKa = 10.16EE436 pKa = 4.79LQGKK440 pKa = 8.03WYY442 pKa = 10.12NVYY445 pKa = 9.69PSPIAVPDD453 pKa = 4.85PIVLNPNRR461 pKa = 11.84EE462 pKa = 4.08RR463 pKa = 11.84EE464 pKa = 4.21WEE466 pKa = 4.3YY467 pKa = 11.26KK468 pKa = 10.38SPNHH472 pKa = 6.2LSKK475 pKa = 10.98SGIYY479 pKa = 8.95SQEE482 pKa = 4.39SIEE485 pKa = 4.32SWQQSLMLASSQSALTKK502 pKa = 10.21IITSKK507 pKa = 11.04FIGKK511 pKa = 9.79DD512 pKa = 2.68IDD514 pKa = 3.92SQGSSIHH521 pKa = 6.11TYY523 pKa = 9.68IDD525 pKa = 3.56EE526 pKa = 6.07DD527 pKa = 3.76IMAHH531 pKa = 5.21ITTNFFVKK539 pKa = 10.26AWGVFSNFGQIMSGLLGFIIIGKK562 pKa = 8.69LFKK565 pKa = 10.81LVVDD569 pKa = 4.96SIMNGIALYY578 pKa = 9.88RR579 pKa = 11.84VYY581 pKa = 10.78GFSIALMGCIWDD593 pKa = 4.03NVAHH597 pKa = 6.62LLISFRR603 pKa = 11.84KK604 pKa = 9.19KK605 pKa = 10.31DD606 pKa = 3.76KK607 pKa = 11.17KK608 pKa = 9.1PTAPSKK614 pKa = 11.13EE615 pKa = 4.21DD616 pKa = 3.33LCDD619 pKa = 3.81NQCEE623 pKa = 4.21PPAAQNPLLTQSTPVSMYY641 pKa = 10.09PNVRR645 pKa = 11.84SMTSHH650 pKa = 7.0EE651 pKa = 4.14LQQ653 pKa = 3.72
MM1 pKa = 7.29YY2 pKa = 9.65IKK4 pKa = 10.58KK5 pKa = 9.84IFWFISLTLCMLEE18 pKa = 3.65ISGLIGFDD26 pKa = 3.8CKK28 pKa = 10.91HH29 pKa = 6.67KK30 pKa = 10.56DD31 pKa = 3.43VEE33 pKa = 4.39YY34 pKa = 10.78SAVSVKK40 pKa = 9.8QVAEE44 pKa = 3.98CDD46 pKa = 3.52EE47 pKa = 4.55VKK49 pKa = 10.54PAVHH53 pKa = 5.57QSKK56 pKa = 10.21MNIQLLQKK64 pKa = 10.21RR65 pKa = 11.84RR66 pKa = 11.84LSKK69 pKa = 10.66VPYY72 pKa = 9.13ISCYY76 pKa = 6.35THH78 pKa = 6.73ANILITHH85 pKa = 7.1CGMHH89 pKa = 5.47SHH91 pKa = 6.8ASVVAGGLSQRR102 pKa = 11.84IEE104 pKa = 4.06QYY106 pKa = 10.28TSEE109 pKa = 4.08TCRR112 pKa = 11.84RR113 pKa = 11.84AFATKK118 pKa = 9.4TVILFGTPIDD128 pKa = 3.53INSFNTTITGSIIPYY143 pKa = 10.01GSVINNDD150 pKa = 3.23ASCIGTSFAYY160 pKa = 10.47NGNHH164 pKa = 4.79YY165 pKa = 9.41RR166 pKa = 11.84ASVMQVSYY174 pKa = 10.77EE175 pKa = 4.19IILKK179 pKa = 8.85TGYY182 pKa = 8.36GTYY185 pKa = 10.47DD186 pKa = 3.28SEE188 pKa = 4.38TGIFNTGLGSYY199 pKa = 10.52GDD201 pKa = 3.84YY202 pKa = 11.03KK203 pKa = 11.06EE204 pKa = 5.59GVLLDD209 pKa = 3.87RR210 pKa = 11.84NFGYY214 pKa = 10.15VFWKK218 pKa = 9.8TSDD221 pKa = 4.18NEE223 pKa = 4.39DD224 pKa = 3.81CSSSTSYY231 pKa = 11.04LVLYY235 pKa = 8.17EE236 pKa = 4.6GPAEE240 pKa = 4.09KK241 pKa = 10.4LVSEE245 pKa = 4.47RR246 pKa = 11.84DD247 pKa = 3.3NRR249 pKa = 11.84SIIVVEE255 pKa = 4.14QQSTTFSLQLRR266 pKa = 11.84EE267 pKa = 4.04PTLLCSQHH275 pKa = 6.36AFNTEE280 pKa = 3.35HH281 pKa = 7.08DD282 pKa = 3.72QLKK285 pKa = 10.54IITSNNRR292 pKa = 11.84MFFFKK297 pKa = 10.61RR298 pKa = 11.84IADD301 pKa = 3.73SLDD304 pKa = 3.43INLSTYY310 pKa = 10.55INAKK314 pKa = 8.1FVYY317 pKa = 9.78VEE319 pKa = 3.55RR320 pKa = 11.84HH321 pKa = 4.48LRR323 pKa = 11.84ANIEE327 pKa = 4.05TLYY330 pKa = 10.44EE331 pKa = 3.87DD332 pKa = 3.63VMKK335 pKa = 10.35FRR337 pKa = 11.84CRR339 pKa = 11.84VTRR342 pKa = 11.84QTLTNLLSLASIDD355 pKa = 3.72EE356 pKa = 4.49DD357 pKa = 3.75EE358 pKa = 4.31FAYY361 pKa = 10.29AYY363 pKa = 8.77MGRR366 pKa = 11.84PGYY369 pKa = 9.38TSVKK373 pKa = 9.14RR374 pKa = 11.84GEE376 pKa = 3.79VAYY379 pKa = 10.31LIKK382 pKa = 10.62CVPTYY387 pKa = 9.59ITLQSSTRR395 pKa = 11.84CYY397 pKa = 8.83NDD399 pKa = 3.06VVISHH404 pKa = 5.84NNKK407 pKa = 8.77TKK409 pKa = 10.59FLASKK414 pKa = 9.43TNIIQDD420 pKa = 3.41FSEE423 pKa = 4.29EE424 pKa = 4.43VEE426 pKa = 4.45CSQLTPVLYY435 pKa = 10.16EE436 pKa = 4.79LQGKK440 pKa = 8.03WYY442 pKa = 10.12NVYY445 pKa = 9.69PSPIAVPDD453 pKa = 4.85PIVLNPNRR461 pKa = 11.84EE462 pKa = 4.08RR463 pKa = 11.84EE464 pKa = 4.21WEE466 pKa = 4.3YY467 pKa = 11.26KK468 pKa = 10.38SPNHH472 pKa = 6.2LSKK475 pKa = 10.98SGIYY479 pKa = 8.95SQEE482 pKa = 4.39SIEE485 pKa = 4.32SWQQSLMLASSQSALTKK502 pKa = 10.21IITSKK507 pKa = 11.04FIGKK511 pKa = 9.79DD512 pKa = 2.68IDD514 pKa = 3.92SQGSSIHH521 pKa = 6.11TYY523 pKa = 9.68IDD525 pKa = 3.56EE526 pKa = 6.07DD527 pKa = 3.76IMAHH531 pKa = 5.21ITTNFFVKK539 pKa = 10.26AWGVFSNFGQIMSGLLGFIIIGKK562 pKa = 8.69LFKK565 pKa = 10.81LVVDD569 pKa = 4.96SIMNGIALYY578 pKa = 9.88RR579 pKa = 11.84VYY581 pKa = 10.78GFSIALMGCIWDD593 pKa = 4.03NVAHH597 pKa = 6.62LLISFRR603 pKa = 11.84KK604 pKa = 9.19KK605 pKa = 10.31DD606 pKa = 3.76KK607 pKa = 11.17KK608 pKa = 9.1PTAPSKK614 pKa = 11.13EE615 pKa = 4.21DD616 pKa = 3.33LCDD619 pKa = 3.81NQCEE623 pKa = 4.21PPAAQNPLLTQSTPVSMYY641 pKa = 10.09PNVRR645 pKa = 11.84SMTSHH650 pKa = 7.0EE651 pKa = 4.14LQQ653 pKa = 3.72
Molecular weight: 73.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMU3|A0A1L3KMU3_9VIRU Uncharacterized protein OS=Hubei odonate virus 11 OX=1922992 PE=4 SV=1
MM1 pKa = 7.31QNPAMEE7 pKa = 4.46VDD9 pKa = 3.68NVRR12 pKa = 11.84PGPSGAQRR20 pKa = 11.84PVVPEE25 pKa = 3.87HH26 pKa = 6.59QILTLGVRR34 pKa = 11.84EE35 pKa = 3.97PRR37 pKa = 11.84YY38 pKa = 10.68AEE40 pKa = 3.71IAALAGKK47 pKa = 8.72PDD49 pKa = 3.32SGFRR53 pKa = 11.84TSPSFSQQLFHH64 pKa = 7.54NLVTPNIMYY73 pKa = 10.44RR74 pKa = 11.84GFLLTHH80 pKa = 6.56TSANFPRR87 pKa = 11.84VMYY90 pKa = 8.55QTGAYY95 pKa = 9.48LAGAVVAIGFAKK107 pKa = 10.02MKK109 pKa = 9.7VPQSATVDD117 pKa = 4.1DD118 pKa = 3.96IKK120 pKa = 11.23LRR122 pKa = 11.84IIGEE126 pKa = 4.07VTAINIAPEE135 pKa = 4.36LLLQTQEE142 pKa = 3.68ADD144 pKa = 3.47ALFLEE149 pKa = 4.44IMNFILRR156 pKa = 11.84SDD158 pKa = 3.56PAIFRR163 pKa = 11.84GGLEE167 pKa = 3.97YY168 pKa = 10.58TSDD171 pKa = 4.02EE172 pKa = 4.34FVSAYY177 pKa = 10.43SDD179 pKa = 3.42PRR181 pKa = 11.84NVDD184 pKa = 3.25VYY186 pKa = 10.97PIGAGFKK193 pKa = 10.34IPLPWSNDD201 pKa = 2.7VEE203 pKa = 4.29LRR205 pKa = 11.84QNSMADD211 pKa = 3.2ATRR214 pKa = 11.84ICLWLTTKK222 pKa = 10.6LANNYY227 pKa = 8.93KK228 pKa = 9.42NCKK231 pKa = 8.97PEE233 pKa = 4.24LSFSYY238 pKa = 10.61LIAICRR244 pKa = 11.84RR245 pKa = 11.84GSVTTQFLTKK255 pKa = 10.46VSDD258 pKa = 5.05DD259 pKa = 3.42INTEE263 pKa = 3.92LGRR266 pKa = 11.84TVSFDD271 pKa = 3.18EE272 pKa = 4.09DD273 pKa = 4.78LIRR276 pKa = 11.84KK277 pKa = 9.3IYY279 pKa = 9.91RR280 pKa = 11.84IYY282 pKa = 11.03GIFINEE288 pKa = 3.68NNAEE292 pKa = 4.07NVLRR296 pKa = 11.84NILTWIPEE304 pKa = 3.71EE305 pKa = 4.13GLRR308 pKa = 11.84LRR310 pKa = 11.84LMLQHH315 pKa = 7.02AEE317 pKa = 4.22GSGLTPHH324 pKa = 5.73YY325 pKa = 8.92TVKK328 pKa = 10.48EE329 pKa = 3.99ALLKK333 pKa = 11.19YY334 pKa = 10.19PDD336 pKa = 4.34FPWGRR341 pKa = 11.84VDD343 pKa = 4.25GLLSGEE349 pKa = 3.6IRR351 pKa = 11.84SFIRR355 pKa = 11.84AWALIKK361 pKa = 11.01GNLYY365 pKa = 10.84YY366 pKa = 10.83GFKK369 pKa = 10.09RR370 pKa = 11.84DD371 pKa = 3.54LKK373 pKa = 9.54EE374 pKa = 4.59ASSQSFKK381 pKa = 11.12GLAYY385 pKa = 9.89VAKK388 pKa = 10.01EE389 pKa = 4.06LLIKK393 pKa = 10.59AAGKK397 pKa = 10.59GGLKK401 pKa = 10.08AYY403 pKa = 10.11RR404 pKa = 11.84GWTRR408 pKa = 11.84TPPSKK413 pKa = 10.9GLLDD417 pKa = 4.68EE418 pKa = 5.67IIEE421 pKa = 4.42NYY423 pKa = 8.62LASMGEE429 pKa = 3.73EE430 pKa = 3.8AAALDD435 pKa = 4.1QTTLDD440 pKa = 3.41QLRR443 pKa = 11.84FIANSTRR450 pKa = 3.22
MM1 pKa = 7.31QNPAMEE7 pKa = 4.46VDD9 pKa = 3.68NVRR12 pKa = 11.84PGPSGAQRR20 pKa = 11.84PVVPEE25 pKa = 3.87HH26 pKa = 6.59QILTLGVRR34 pKa = 11.84EE35 pKa = 3.97PRR37 pKa = 11.84YY38 pKa = 10.68AEE40 pKa = 3.71IAALAGKK47 pKa = 8.72PDD49 pKa = 3.32SGFRR53 pKa = 11.84TSPSFSQQLFHH64 pKa = 7.54NLVTPNIMYY73 pKa = 10.44RR74 pKa = 11.84GFLLTHH80 pKa = 6.56TSANFPRR87 pKa = 11.84VMYY90 pKa = 8.55QTGAYY95 pKa = 9.48LAGAVVAIGFAKK107 pKa = 10.02MKK109 pKa = 9.7VPQSATVDD117 pKa = 4.1DD118 pKa = 3.96IKK120 pKa = 11.23LRR122 pKa = 11.84IIGEE126 pKa = 4.07VTAINIAPEE135 pKa = 4.36LLLQTQEE142 pKa = 3.68ADD144 pKa = 3.47ALFLEE149 pKa = 4.44IMNFILRR156 pKa = 11.84SDD158 pKa = 3.56PAIFRR163 pKa = 11.84GGLEE167 pKa = 3.97YY168 pKa = 10.58TSDD171 pKa = 4.02EE172 pKa = 4.34FVSAYY177 pKa = 10.43SDD179 pKa = 3.42PRR181 pKa = 11.84NVDD184 pKa = 3.25VYY186 pKa = 10.97PIGAGFKK193 pKa = 10.34IPLPWSNDD201 pKa = 2.7VEE203 pKa = 4.29LRR205 pKa = 11.84QNSMADD211 pKa = 3.2ATRR214 pKa = 11.84ICLWLTTKK222 pKa = 10.6LANNYY227 pKa = 8.93KK228 pKa = 9.42NCKK231 pKa = 8.97PEE233 pKa = 4.24LSFSYY238 pKa = 10.61LIAICRR244 pKa = 11.84RR245 pKa = 11.84GSVTTQFLTKK255 pKa = 10.46VSDD258 pKa = 5.05DD259 pKa = 3.42INTEE263 pKa = 3.92LGRR266 pKa = 11.84TVSFDD271 pKa = 3.18EE272 pKa = 4.09DD273 pKa = 4.78LIRR276 pKa = 11.84KK277 pKa = 9.3IYY279 pKa = 9.91RR280 pKa = 11.84IYY282 pKa = 11.03GIFINEE288 pKa = 3.68NNAEE292 pKa = 4.07NVLRR296 pKa = 11.84NILTWIPEE304 pKa = 3.71EE305 pKa = 4.13GLRR308 pKa = 11.84LRR310 pKa = 11.84LMLQHH315 pKa = 7.02AEE317 pKa = 4.22GSGLTPHH324 pKa = 5.73YY325 pKa = 8.92TVKK328 pKa = 10.48EE329 pKa = 3.99ALLKK333 pKa = 11.19YY334 pKa = 10.19PDD336 pKa = 4.34FPWGRR341 pKa = 11.84VDD343 pKa = 4.25GLLSGEE349 pKa = 3.6IRR351 pKa = 11.84SFIRR355 pKa = 11.84AWALIKK361 pKa = 11.01GNLYY365 pKa = 10.84YY366 pKa = 10.83GFKK369 pKa = 10.09RR370 pKa = 11.84DD371 pKa = 3.54LKK373 pKa = 9.54EE374 pKa = 4.59ASSQSFKK381 pKa = 11.12GLAYY385 pKa = 9.89VAKK388 pKa = 10.01EE389 pKa = 4.06LLIKK393 pKa = 10.59AAGKK397 pKa = 10.59GGLKK401 pKa = 10.08AYY403 pKa = 10.11RR404 pKa = 11.84GWTRR408 pKa = 11.84TPPSKK413 pKa = 10.9GLLDD417 pKa = 4.68EE418 pKa = 5.67IIEE421 pKa = 4.42NYY423 pKa = 8.62LASMGEE429 pKa = 3.73EE430 pKa = 3.8AAALDD435 pKa = 4.1QTTLDD440 pKa = 3.41QLRR443 pKa = 11.84FIANSTRR450 pKa = 3.22
Molecular weight: 50.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3271 |
450 |
2168 |
1090.3 |
124.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.044 ± 1.021 | 2.66 ± 0.574 |
4.983 ± 0.246 | 5.319 ± 0.191 |
4.127 ± 0.169 | 4.953 ± 0.627 |
2.293 ± 0.302 | 7.49 ± 0.372 |
6.023 ± 0.436 | 10.639 ± 0.887 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.14 ± 0.055 | 5.105 ± 0.056 |
3.944 ± 0.352 | 3.76 ± 0.141 |
4.769 ± 0.45 | 8.224 ± 0.755 |
6.084 ± 0.222 | 6.389 ± 0.364 |
1.192 ± 0.046 | 4.861 ± 0.217 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
