
Amphibalanus amphitrite (Striped barnacle) (Balanus amphitrite)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Crustacea; Multicrustacea; Hexanauplia; Thecostraca; Cirripedia; Thoracica; Sessilia; Balanidae; Amphibalanus
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
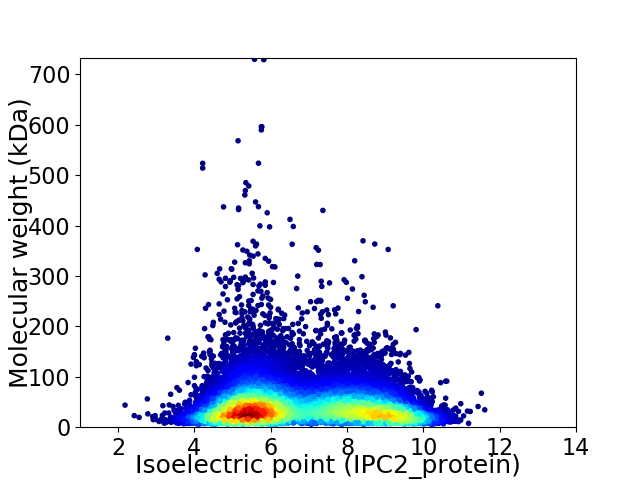
Virtual 2D-PAGE plot for 26159 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6A4WIG3|A0A6A4WIG3_AMPAM Kelch-like protein 20 OS=Amphibalanus amphitrite OX=1232801 GN=Klhl20 PE=4 SV=1
MM1 pKa = 7.37RR2 pKa = 11.84RR3 pKa = 11.84CVGRR7 pKa = 11.84TLRR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 8.3GTDD15 pKa = 2.91TTLEE19 pKa = 4.7DD20 pKa = 4.29PSQSDD25 pKa = 3.66DD26 pKa = 3.25VTDD29 pKa = 4.51RR30 pKa = 11.84VTEE33 pKa = 4.16DD34 pKa = 3.03LTDD37 pKa = 3.88GVVSGSVSEE46 pKa = 4.39QAWDD50 pKa = 4.06PYY52 pKa = 10.32QASSDD57 pKa = 3.6SDD59 pKa = 4.04SVSEE63 pKa = 4.26QAWDD67 pKa = 4.06PYY69 pKa = 10.33QASSDD74 pKa = 3.72SGSVSEE80 pKa = 4.75QAWDD84 pKa = 3.99PYY86 pKa = 10.12QASSASGSVSEE97 pKa = 4.66QAWDD101 pKa = 3.99PYY103 pKa = 10.12QASSASGSVSEE114 pKa = 4.66QAWDD118 pKa = 3.99PYY120 pKa = 10.12QASSASGSVSGQAWDD135 pKa = 4.42PYY137 pKa = 10.12QASSDD142 pKa = 3.72SGSVSEE148 pKa = 4.75QAWDD152 pKa = 3.99PYY154 pKa = 10.12QASSASGSVSEE165 pKa = 4.66QAWDD169 pKa = 4.06PYY171 pKa = 10.32QASSDD176 pKa = 3.6SDD178 pKa = 4.04SVSEE182 pKa = 4.26QAWDD186 pKa = 4.06PYY188 pKa = 10.33QASSDD193 pKa = 3.72SGSVSEE199 pKa = 4.75QAWDD203 pKa = 3.99PYY205 pKa = 10.12QASSASGSVSEE216 pKa = 4.66QAWDD220 pKa = 3.99PYY222 pKa = 10.12QASSASGSVSEE233 pKa = 4.66QAWDD237 pKa = 3.99PYY239 pKa = 10.12QASSASGSVSGQAWDD254 pKa = 4.42PYY256 pKa = 10.12QASSDD261 pKa = 3.72SGSVSEE267 pKa = 4.75QAWDD271 pKa = 3.99PYY273 pKa = 10.03QASSASDD280 pKa = 3.58SVSEE284 pKa = 4.23QAWDD288 pKa = 3.99PYY290 pKa = 10.03QASSASDD297 pKa = 3.58SVSEE301 pKa = 4.23QAWDD305 pKa = 4.06PYY307 pKa = 10.33QASSDD312 pKa = 3.72SGSVSEE318 pKa = 4.75QAWDD322 pKa = 4.06PYY324 pKa = 10.32QASSDD329 pKa = 3.89SDD331 pKa = 3.53SQEE334 pKa = 3.84YY335 pKa = 10.55KK336 pKa = 10.42EE337 pKa = 4.12VSEE340 pKa = 4.92PGSEE344 pKa = 3.91PAVDD348 pKa = 3.83PEE350 pKa = 4.11TLRR353 pKa = 11.84RR354 pKa = 11.84FLDD357 pKa = 3.4TDD359 pKa = 3.81DD360 pKa = 3.65YY361 pKa = 11.83RR362 pKa = 11.84NFIDD366 pKa = 4.96SLSDD370 pKa = 3.46CTSSQVSGAPARR382 pKa = 11.84PPPPRR387 pKa = 11.84RR388 pKa = 11.84QRR390 pKa = 11.84RR391 pKa = 11.84KK392 pKa = 9.88LRR394 pKa = 11.84AAASDD399 pKa = 4.01SVDD402 pKa = 3.24SDD404 pKa = 3.49TDD406 pKa = 3.52GGDD409 pKa = 3.24EE410 pKa = 3.9VAAALVEE417 pKa = 4.38SRR419 pKa = 11.84RR420 pKa = 11.84RR421 pKa = 11.84LQLARR426 pKa = 11.84TEE428 pKa = 4.21LSRR431 pKa = 11.84RR432 pKa = 11.84AHH434 pKa = 6.1QPDD437 pKa = 5.33PICVSNGYY445 pKa = 9.1RR446 pKa = 11.84VPAGPQYY453 pKa = 10.27RR454 pKa = 11.84VPAGPHH460 pKa = 5.35LCVV463 pKa = 3.59
MM1 pKa = 7.37RR2 pKa = 11.84RR3 pKa = 11.84CVGRR7 pKa = 11.84TLRR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 8.3GTDD15 pKa = 2.91TTLEE19 pKa = 4.7DD20 pKa = 4.29PSQSDD25 pKa = 3.66DD26 pKa = 3.25VTDD29 pKa = 4.51RR30 pKa = 11.84VTEE33 pKa = 4.16DD34 pKa = 3.03LTDD37 pKa = 3.88GVVSGSVSEE46 pKa = 4.39QAWDD50 pKa = 4.06PYY52 pKa = 10.32QASSDD57 pKa = 3.6SDD59 pKa = 4.04SVSEE63 pKa = 4.26QAWDD67 pKa = 4.06PYY69 pKa = 10.33QASSDD74 pKa = 3.72SGSVSEE80 pKa = 4.75QAWDD84 pKa = 3.99PYY86 pKa = 10.12QASSASGSVSEE97 pKa = 4.66QAWDD101 pKa = 3.99PYY103 pKa = 10.12QASSASGSVSEE114 pKa = 4.66QAWDD118 pKa = 3.99PYY120 pKa = 10.12QASSASGSVSGQAWDD135 pKa = 4.42PYY137 pKa = 10.12QASSDD142 pKa = 3.72SGSVSEE148 pKa = 4.75QAWDD152 pKa = 3.99PYY154 pKa = 10.12QASSASGSVSEE165 pKa = 4.66QAWDD169 pKa = 4.06PYY171 pKa = 10.32QASSDD176 pKa = 3.6SDD178 pKa = 4.04SVSEE182 pKa = 4.26QAWDD186 pKa = 4.06PYY188 pKa = 10.33QASSDD193 pKa = 3.72SGSVSEE199 pKa = 4.75QAWDD203 pKa = 3.99PYY205 pKa = 10.12QASSASGSVSEE216 pKa = 4.66QAWDD220 pKa = 3.99PYY222 pKa = 10.12QASSASGSVSEE233 pKa = 4.66QAWDD237 pKa = 3.99PYY239 pKa = 10.12QASSASGSVSGQAWDD254 pKa = 4.42PYY256 pKa = 10.12QASSDD261 pKa = 3.72SGSVSEE267 pKa = 4.75QAWDD271 pKa = 3.99PYY273 pKa = 10.03QASSASDD280 pKa = 3.58SVSEE284 pKa = 4.23QAWDD288 pKa = 3.99PYY290 pKa = 10.03QASSASDD297 pKa = 3.58SVSEE301 pKa = 4.23QAWDD305 pKa = 4.06PYY307 pKa = 10.33QASSDD312 pKa = 3.72SGSVSEE318 pKa = 4.75QAWDD322 pKa = 4.06PYY324 pKa = 10.32QASSDD329 pKa = 3.89SDD331 pKa = 3.53SQEE334 pKa = 3.84YY335 pKa = 10.55KK336 pKa = 10.42EE337 pKa = 4.12VSEE340 pKa = 4.92PGSEE344 pKa = 3.91PAVDD348 pKa = 3.83PEE350 pKa = 4.11TLRR353 pKa = 11.84RR354 pKa = 11.84FLDD357 pKa = 3.4TDD359 pKa = 3.81DD360 pKa = 3.65YY361 pKa = 11.83RR362 pKa = 11.84NFIDD366 pKa = 4.96SLSDD370 pKa = 3.46CTSSQVSGAPARR382 pKa = 11.84PPPPRR387 pKa = 11.84RR388 pKa = 11.84QRR390 pKa = 11.84RR391 pKa = 11.84KK392 pKa = 9.88LRR394 pKa = 11.84AAASDD399 pKa = 4.01SVDD402 pKa = 3.24SDD404 pKa = 3.49TDD406 pKa = 3.52GGDD409 pKa = 3.24EE410 pKa = 3.9VAAALVEE417 pKa = 4.38SRR419 pKa = 11.84RR420 pKa = 11.84RR421 pKa = 11.84LQLARR426 pKa = 11.84TEE428 pKa = 4.21LSRR431 pKa = 11.84RR432 pKa = 11.84AHH434 pKa = 6.1QPDD437 pKa = 5.33PICVSNGYY445 pKa = 9.1RR446 pKa = 11.84VPAGPQYY453 pKa = 10.27RR454 pKa = 11.84VPAGPHH460 pKa = 5.35LCVV463 pKa = 3.59
Molecular weight: 49.45 kDa
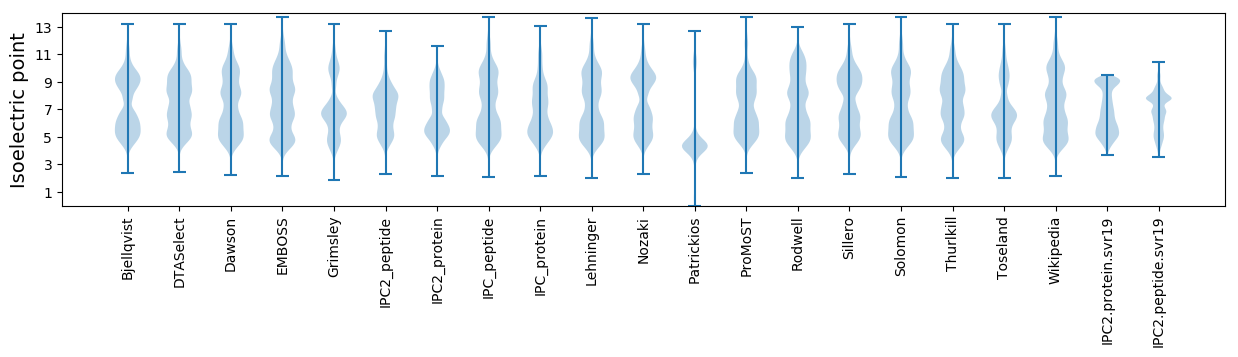
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6A4X3Q4|A0A6A4X3Q4_AMPAM Uncharacterized protein OS=Amphibalanus amphitrite OX=1232801 GN=FJT64_019337 PE=4 SV=1
MM1 pKa = 7.23IRR3 pKa = 11.84WPQTALVRR11 pKa = 11.84QTALVRR17 pKa = 11.84QTALVRR23 pKa = 11.84RR24 pKa = 11.84TTLVRR29 pKa = 11.84QTALVRR35 pKa = 11.84RR36 pKa = 11.84TALVRR41 pKa = 11.84RR42 pKa = 11.84TALVRR47 pKa = 11.84QTALVRR53 pKa = 11.84RR54 pKa = 11.84TALVRR59 pKa = 11.84RR60 pKa = 11.84TALVRR65 pKa = 11.84QTALVRR71 pKa = 11.84QTALVRR77 pKa = 11.84QTVLVRR83 pKa = 11.84QTALVRR89 pKa = 11.84QTVLVRR95 pKa = 11.84QTALVRR101 pKa = 11.84QTALVRR107 pKa = 11.84RR108 pKa = 11.84TALVRR113 pKa = 11.84QTALVRR119 pKa = 11.84RR120 pKa = 11.84TALVRR125 pKa = 11.84RR126 pKa = 11.84TALVRR131 pKa = 11.84RR132 pKa = 11.84TALVRR137 pKa = 11.84QTALVRR143 pKa = 11.84QTALVRR149 pKa = 11.84QTALVRR155 pKa = 11.84QTALVRR161 pKa = 11.84QTALVRR167 pKa = 11.84RR168 pKa = 11.84TALVRR173 pKa = 11.84QTALVRR179 pKa = 11.84QTALVRR185 pKa = 11.84RR186 pKa = 11.84TALVRR191 pKa = 11.84QTALVRR197 pKa = 11.84RR198 pKa = 11.84TALVRR203 pKa = 11.84RR204 pKa = 11.84TALVRR209 pKa = 11.84RR210 pKa = 11.84TALVRR215 pKa = 11.84RR216 pKa = 11.84TALVRR221 pKa = 11.84RR222 pKa = 11.84TALVRR227 pKa = 11.84QTALMWMLATLVALDD242 pKa = 4.72PGHH245 pKa = 6.57SGWCFHH251 pKa = 7.08PLLSRR256 pKa = 11.84QMASEE261 pKa = 3.8ILVPDD266 pKa = 4.5RR267 pKa = 11.84MAGWKK272 pKa = 10.23VNN274 pKa = 3.41
MM1 pKa = 7.23IRR3 pKa = 11.84WPQTALVRR11 pKa = 11.84QTALVRR17 pKa = 11.84QTALVRR23 pKa = 11.84RR24 pKa = 11.84TTLVRR29 pKa = 11.84QTALVRR35 pKa = 11.84RR36 pKa = 11.84TALVRR41 pKa = 11.84RR42 pKa = 11.84TALVRR47 pKa = 11.84QTALVRR53 pKa = 11.84RR54 pKa = 11.84TALVRR59 pKa = 11.84RR60 pKa = 11.84TALVRR65 pKa = 11.84QTALVRR71 pKa = 11.84QTALVRR77 pKa = 11.84QTVLVRR83 pKa = 11.84QTALVRR89 pKa = 11.84QTVLVRR95 pKa = 11.84QTALVRR101 pKa = 11.84QTALVRR107 pKa = 11.84RR108 pKa = 11.84TALVRR113 pKa = 11.84QTALVRR119 pKa = 11.84RR120 pKa = 11.84TALVRR125 pKa = 11.84RR126 pKa = 11.84TALVRR131 pKa = 11.84RR132 pKa = 11.84TALVRR137 pKa = 11.84QTALVRR143 pKa = 11.84QTALVRR149 pKa = 11.84QTALVRR155 pKa = 11.84QTALVRR161 pKa = 11.84QTALVRR167 pKa = 11.84RR168 pKa = 11.84TALVRR173 pKa = 11.84QTALVRR179 pKa = 11.84QTALVRR185 pKa = 11.84RR186 pKa = 11.84TALVRR191 pKa = 11.84QTALVRR197 pKa = 11.84RR198 pKa = 11.84TALVRR203 pKa = 11.84RR204 pKa = 11.84TALVRR209 pKa = 11.84RR210 pKa = 11.84TALVRR215 pKa = 11.84RR216 pKa = 11.84TALVRR221 pKa = 11.84RR222 pKa = 11.84TALVRR227 pKa = 11.84QTALMWMLATLVALDD242 pKa = 4.72PGHH245 pKa = 6.57SGWCFHH251 pKa = 7.08PLLSRR256 pKa = 11.84QMASEE261 pKa = 3.8ILVPDD266 pKa = 4.5RR267 pKa = 11.84MAGWKK272 pKa = 10.23VNN274 pKa = 3.41
Molecular weight: 31.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11229583 |
50 |
8844 |
429.3 |
47.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.954 ± 0.023 | 2.06 ± 0.016 |
5.489 ± 0.012 | 6.304 ± 0.019 |
2.943 ± 0.01 | 7.306 ± 0.019 |
2.41 ± 0.007 | 3.143 ± 0.012 |
3.598 ± 0.016 | 9.582 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.032 ± 0.007 | 2.586 ± 0.01 |
6.753 ± 0.021 | 4.358 ± 0.013 |
7.903 ± 0.017 | 7.435 ± 0.017 |
5.583 ± 0.012 | 6.853 ± 0.016 |
1.3 ± 0.006 | 2.407 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
