
SARS coronavirus PUMC02
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Sarbecovirus; Severe acute respiratory syndrome-related coronavirus
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
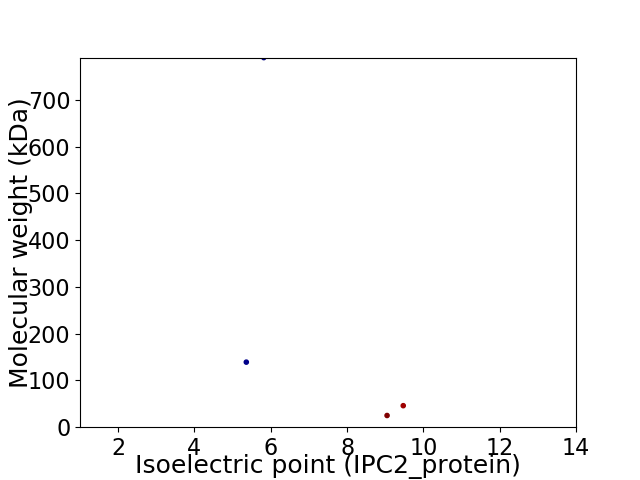
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6UZF5|Q6UZF5_SARS 2'-O-methyltransferase OS=SARS coronavirus PUMC02 OX=253434 PE=3 SV=1
MM1 pKa = 7.88FIFLLFLTLTSGSDD15 pKa = 3.56LDD17 pKa = 3.81RR18 pKa = 11.84CTTFDD23 pKa = 5.11DD24 pKa = 4.16VQAPNYY30 pKa = 6.73TQHH33 pKa = 5.96TSSMRR38 pKa = 11.84GVYY41 pKa = 10.09YY42 pKa = 9.5PDD44 pKa = 3.78EE45 pKa = 4.48IFRR48 pKa = 11.84SDD50 pKa = 3.65TLYY53 pKa = 9.95LTQDD57 pKa = 3.75LFLPFYY63 pKa = 11.15SNVTGFHH70 pKa = 6.93TINHH74 pKa = 5.98TFGNPVIPFKK84 pKa = 11.08DD85 pKa = 3.93GIYY88 pKa = 10.18FAATEE93 pKa = 4.03KK94 pKa = 11.08SNVVRR99 pKa = 11.84GWVFGSTMNNKK110 pKa = 8.76SQSVIIINNSTNVVIRR126 pKa = 11.84ACNFEE131 pKa = 4.97LCDD134 pKa = 3.57NPFFAVSKK142 pKa = 10.56PMGTQTHH149 pKa = 5.42TMIFDD154 pKa = 3.4NAFNCTFEE162 pKa = 4.55YY163 pKa = 10.53ISDD166 pKa = 3.88AFSLDD171 pKa = 3.34VSEE174 pKa = 4.82KK175 pKa = 10.62SGNFKK180 pKa = 10.48HH181 pKa = 6.67LRR183 pKa = 11.84EE184 pKa = 4.1FVFKK188 pKa = 11.13NKK190 pKa = 10.4DD191 pKa = 3.05GFLYY195 pKa = 10.4VYY197 pKa = 10.28KK198 pKa = 10.23GYY200 pKa = 10.7QPIDD204 pKa = 3.59VVRR207 pKa = 11.84DD208 pKa = 3.53LPSGFNTLKK217 pKa = 10.74PIFKK221 pKa = 10.35LPLGINITNFRR232 pKa = 11.84AILTAFSPAQDD243 pKa = 2.57IWGTSAAAYY252 pKa = 9.13FVGYY256 pKa = 9.98LKK258 pKa = 9.58PTTFMLKK265 pKa = 9.68YY266 pKa = 10.06DD267 pKa = 4.12EE268 pKa = 5.37NGTITDD274 pKa = 5.15AVDD277 pKa = 3.92CSQNPLAEE285 pKa = 4.52LKK287 pKa = 10.85CSVKK291 pKa = 10.59SFEE294 pKa = 4.13IDD296 pKa = 2.72KK297 pKa = 11.25GIYY300 pKa = 6.46QTSNFRR306 pKa = 11.84VVPSGDD312 pKa = 3.08VVRR315 pKa = 11.84FPNITNLCPFGEE327 pKa = 4.44VFNATKK333 pKa = 10.41FPSVYY338 pKa = 9.44AWEE341 pKa = 4.09RR342 pKa = 11.84KK343 pKa = 9.67KK344 pKa = 10.83ISNCVADD351 pKa = 4.37YY352 pKa = 11.15SVLYY356 pKa = 10.67NSTFFSTFKK365 pKa = 10.6CYY367 pKa = 10.02GVSATKK373 pKa = 10.88LNDD376 pKa = 3.32LCFSNVYY383 pKa = 10.62ADD385 pKa = 3.79SFVVKK390 pKa = 10.43GDD392 pKa = 3.78DD393 pKa = 3.46VRR395 pKa = 11.84QIAPGQTGVIADD407 pKa = 4.17YY408 pKa = 10.79NYY410 pKa = 11.15KK411 pKa = 10.75LPDD414 pKa = 3.8DD415 pKa = 4.41FMGCVLAWNTRR426 pKa = 11.84NIDD429 pKa = 3.36ATSTGNYY436 pKa = 7.96NYY438 pKa = 10.14KK439 pKa = 10.12YY440 pKa = 10.29RR441 pKa = 11.84YY442 pKa = 8.78LRR444 pKa = 11.84HH445 pKa = 5.69GKK447 pKa = 9.14LRR449 pKa = 11.84PFEE452 pKa = 4.59RR453 pKa = 11.84DD454 pKa = 2.59ISNVPFSPDD463 pKa = 3.26GKK465 pKa = 9.6PCTPPALNCYY475 pKa = 8.9WPLNDD480 pKa = 3.46YY481 pKa = 11.13GFYY484 pKa = 7.22TTTGIGYY491 pKa = 8.17QPYY494 pKa = 9.17RR495 pKa = 11.84VVVLSFEE502 pKa = 4.61LLNAPATVCGPKK514 pKa = 10.44LSTDD518 pKa = 4.57LIKK521 pKa = 10.91NQCVNFNFNGLTGTGVLTPSSKK543 pKa = 10.22RR544 pKa = 11.84FQPFQQFGRR553 pKa = 11.84DD554 pKa = 3.25VSDD557 pKa = 3.54FTDD560 pKa = 3.29SVRR563 pKa = 11.84DD564 pKa = 3.75PKK566 pKa = 10.71TSEE569 pKa = 3.88ILDD572 pKa = 3.48ISPCSFGGVSVITPGTNASSEE593 pKa = 4.47VAVLYY598 pKa = 10.73QDD600 pKa = 4.28VNCTDD605 pKa = 3.12VSTAIHH611 pKa = 7.03ADD613 pKa = 3.02QLTPAWRR620 pKa = 11.84IYY622 pKa = 9.61STGNNVFQTQAGCLIGAEE640 pKa = 4.41HH641 pKa = 7.11VDD643 pKa = 3.32TSYY646 pKa = 11.61EE647 pKa = 3.86CDD649 pKa = 3.16IPIGAGICASYY660 pKa = 8.45HH661 pKa = 4.57TVSLLRR667 pKa = 11.84STSQKK672 pKa = 10.89SIVAYY677 pKa = 9.62TMSLGADD684 pKa = 3.28SSIAYY689 pKa = 10.1SNNTIAIPTNFSISITTEE707 pKa = 3.64VMPVSMAKK715 pKa = 9.66TSVDD719 pKa = 3.3CNMYY723 pKa = 9.84ICGDD727 pKa = 3.6STEE730 pKa = 4.39CANLLLQYY738 pKa = 11.2GSFCTQLNRR747 pKa = 11.84ALSGIAAEE755 pKa = 4.17QDD757 pKa = 3.08RR758 pKa = 11.84NTRR761 pKa = 11.84EE762 pKa = 3.35VFAQVKK768 pKa = 9.02QMYY771 pKa = 7.48KK772 pKa = 9.69TPTLKK777 pKa = 10.84YY778 pKa = 9.33FGGFNFSQILPDD790 pKa = 3.84PLKK793 pKa = 8.1PTKK796 pKa = 10.2RR797 pKa = 11.84SFIEE801 pKa = 4.3DD802 pKa = 3.61LLFNKK807 pKa = 7.99VTLADD812 pKa = 3.57AGFMKK817 pKa = 10.37QYY819 pKa = 10.74GEE821 pKa = 4.31CLGDD825 pKa = 3.25INARR829 pKa = 11.84DD830 pKa = 4.56LICAQKK836 pKa = 10.77FNGLTVLPPLLTDD849 pKa = 4.9DD850 pKa = 4.88MIAAYY855 pKa = 8.09TAALVSGTATAGWTFGAGAALQIPFAMQMAYY886 pKa = 10.08RR887 pKa = 11.84FNGIGVTQNVLYY899 pKa = 10.46EE900 pKa = 4.09NQKK903 pKa = 10.62QIANQFNKK911 pKa = 10.46AISQIQEE918 pKa = 4.19SLTTTSTALGKK929 pKa = 10.37LQDD932 pKa = 3.95VVNQNAQALNTLVKK946 pKa = 10.38QLSSNFGAISSVLNDD961 pKa = 3.17ILSRR965 pKa = 11.84LDD967 pKa = 3.31KK968 pKa = 11.45VEE970 pKa = 5.19AEE972 pKa = 4.28VQIDD976 pKa = 3.56RR977 pKa = 11.84LITGRR982 pKa = 11.84LQSLQTYY989 pKa = 7.4VTQQLIRR996 pKa = 11.84AAEE999 pKa = 3.85IRR1001 pKa = 11.84ASANLAATKK1010 pKa = 9.96MSEE1013 pKa = 4.28CVLGQSKK1020 pKa = 10.12RR1021 pKa = 11.84VDD1023 pKa = 4.0FCGKK1027 pKa = 9.92GYY1029 pKa = 10.57HH1030 pKa = 6.84LMSFPQAAPHH1040 pKa = 5.76GVVFLHH1046 pKa = 5.25VTYY1049 pKa = 10.82VPSQEE1054 pKa = 4.9RR1055 pKa = 11.84NFTTAPAICHH1065 pKa = 5.73EE1066 pKa = 4.45GKK1068 pKa = 10.56AYY1070 pKa = 9.93FPRR1073 pKa = 11.84EE1074 pKa = 3.72GVFVFNGTSWFITQRR1089 pKa = 11.84NFFSPQIITTDD1100 pKa = 3.16NTFVSGNCDD1109 pKa = 3.2VVIGIINNTVYY1120 pKa = 10.97DD1121 pKa = 4.22PLQPEE1126 pKa = 4.19LDD1128 pKa = 3.85SFKK1131 pKa = 11.4EE1132 pKa = 3.91EE1133 pKa = 3.56LDD1135 pKa = 3.63KK1136 pKa = 11.53YY1137 pKa = 10.15FKK1139 pKa = 10.99NHH1141 pKa = 6.01TSPDD1145 pKa = 3.14VDD1147 pKa = 4.4LGDD1150 pKa = 3.31ISGINASVVNIQKK1163 pKa = 10.63EE1164 pKa = 3.92IDD1166 pKa = 3.67RR1167 pKa = 11.84LNEE1170 pKa = 3.5VAKK1173 pKa = 10.61NLNEE1177 pKa = 4.14SLIDD1181 pKa = 3.75LQEE1184 pKa = 4.17LGKK1187 pKa = 10.54YY1188 pKa = 6.98EE1189 pKa = 5.3QYY1191 pKa = 10.78IKK1193 pKa = 10.03WPWYY1197 pKa = 8.39VWLGFIAGLIAIVMVTILLCCMTSCCSCLKK1227 pKa = 10.16GACSCGSCCKK1237 pKa = 10.16FDD1239 pKa = 4.43EE1240 pKa = 5.39DD1241 pKa = 5.38DD1242 pKa = 4.38SEE1244 pKa = 4.36PVLKK1248 pKa = 10.36GVKK1251 pKa = 9.35LHH1253 pKa = 5.3YY1254 pKa = 9.44TT1255 pKa = 3.44
MM1 pKa = 7.88FIFLLFLTLTSGSDD15 pKa = 3.56LDD17 pKa = 3.81RR18 pKa = 11.84CTTFDD23 pKa = 5.11DD24 pKa = 4.16VQAPNYY30 pKa = 6.73TQHH33 pKa = 5.96TSSMRR38 pKa = 11.84GVYY41 pKa = 10.09YY42 pKa = 9.5PDD44 pKa = 3.78EE45 pKa = 4.48IFRR48 pKa = 11.84SDD50 pKa = 3.65TLYY53 pKa = 9.95LTQDD57 pKa = 3.75LFLPFYY63 pKa = 11.15SNVTGFHH70 pKa = 6.93TINHH74 pKa = 5.98TFGNPVIPFKK84 pKa = 11.08DD85 pKa = 3.93GIYY88 pKa = 10.18FAATEE93 pKa = 4.03KK94 pKa = 11.08SNVVRR99 pKa = 11.84GWVFGSTMNNKK110 pKa = 8.76SQSVIIINNSTNVVIRR126 pKa = 11.84ACNFEE131 pKa = 4.97LCDD134 pKa = 3.57NPFFAVSKK142 pKa = 10.56PMGTQTHH149 pKa = 5.42TMIFDD154 pKa = 3.4NAFNCTFEE162 pKa = 4.55YY163 pKa = 10.53ISDD166 pKa = 3.88AFSLDD171 pKa = 3.34VSEE174 pKa = 4.82KK175 pKa = 10.62SGNFKK180 pKa = 10.48HH181 pKa = 6.67LRR183 pKa = 11.84EE184 pKa = 4.1FVFKK188 pKa = 11.13NKK190 pKa = 10.4DD191 pKa = 3.05GFLYY195 pKa = 10.4VYY197 pKa = 10.28KK198 pKa = 10.23GYY200 pKa = 10.7QPIDD204 pKa = 3.59VVRR207 pKa = 11.84DD208 pKa = 3.53LPSGFNTLKK217 pKa = 10.74PIFKK221 pKa = 10.35LPLGINITNFRR232 pKa = 11.84AILTAFSPAQDD243 pKa = 2.57IWGTSAAAYY252 pKa = 9.13FVGYY256 pKa = 9.98LKK258 pKa = 9.58PTTFMLKK265 pKa = 9.68YY266 pKa = 10.06DD267 pKa = 4.12EE268 pKa = 5.37NGTITDD274 pKa = 5.15AVDD277 pKa = 3.92CSQNPLAEE285 pKa = 4.52LKK287 pKa = 10.85CSVKK291 pKa = 10.59SFEE294 pKa = 4.13IDD296 pKa = 2.72KK297 pKa = 11.25GIYY300 pKa = 6.46QTSNFRR306 pKa = 11.84VVPSGDD312 pKa = 3.08VVRR315 pKa = 11.84FPNITNLCPFGEE327 pKa = 4.44VFNATKK333 pKa = 10.41FPSVYY338 pKa = 9.44AWEE341 pKa = 4.09RR342 pKa = 11.84KK343 pKa = 9.67KK344 pKa = 10.83ISNCVADD351 pKa = 4.37YY352 pKa = 11.15SVLYY356 pKa = 10.67NSTFFSTFKK365 pKa = 10.6CYY367 pKa = 10.02GVSATKK373 pKa = 10.88LNDD376 pKa = 3.32LCFSNVYY383 pKa = 10.62ADD385 pKa = 3.79SFVVKK390 pKa = 10.43GDD392 pKa = 3.78DD393 pKa = 3.46VRR395 pKa = 11.84QIAPGQTGVIADD407 pKa = 4.17YY408 pKa = 10.79NYY410 pKa = 11.15KK411 pKa = 10.75LPDD414 pKa = 3.8DD415 pKa = 4.41FMGCVLAWNTRR426 pKa = 11.84NIDD429 pKa = 3.36ATSTGNYY436 pKa = 7.96NYY438 pKa = 10.14KK439 pKa = 10.12YY440 pKa = 10.29RR441 pKa = 11.84YY442 pKa = 8.78LRR444 pKa = 11.84HH445 pKa = 5.69GKK447 pKa = 9.14LRR449 pKa = 11.84PFEE452 pKa = 4.59RR453 pKa = 11.84DD454 pKa = 2.59ISNVPFSPDD463 pKa = 3.26GKK465 pKa = 9.6PCTPPALNCYY475 pKa = 8.9WPLNDD480 pKa = 3.46YY481 pKa = 11.13GFYY484 pKa = 7.22TTTGIGYY491 pKa = 8.17QPYY494 pKa = 9.17RR495 pKa = 11.84VVVLSFEE502 pKa = 4.61LLNAPATVCGPKK514 pKa = 10.44LSTDD518 pKa = 4.57LIKK521 pKa = 10.91NQCVNFNFNGLTGTGVLTPSSKK543 pKa = 10.22RR544 pKa = 11.84FQPFQQFGRR553 pKa = 11.84DD554 pKa = 3.25VSDD557 pKa = 3.54FTDD560 pKa = 3.29SVRR563 pKa = 11.84DD564 pKa = 3.75PKK566 pKa = 10.71TSEE569 pKa = 3.88ILDD572 pKa = 3.48ISPCSFGGVSVITPGTNASSEE593 pKa = 4.47VAVLYY598 pKa = 10.73QDD600 pKa = 4.28VNCTDD605 pKa = 3.12VSTAIHH611 pKa = 7.03ADD613 pKa = 3.02QLTPAWRR620 pKa = 11.84IYY622 pKa = 9.61STGNNVFQTQAGCLIGAEE640 pKa = 4.41HH641 pKa = 7.11VDD643 pKa = 3.32TSYY646 pKa = 11.61EE647 pKa = 3.86CDD649 pKa = 3.16IPIGAGICASYY660 pKa = 8.45HH661 pKa = 4.57TVSLLRR667 pKa = 11.84STSQKK672 pKa = 10.89SIVAYY677 pKa = 9.62TMSLGADD684 pKa = 3.28SSIAYY689 pKa = 10.1SNNTIAIPTNFSISITTEE707 pKa = 3.64VMPVSMAKK715 pKa = 9.66TSVDD719 pKa = 3.3CNMYY723 pKa = 9.84ICGDD727 pKa = 3.6STEE730 pKa = 4.39CANLLLQYY738 pKa = 11.2GSFCTQLNRR747 pKa = 11.84ALSGIAAEE755 pKa = 4.17QDD757 pKa = 3.08RR758 pKa = 11.84NTRR761 pKa = 11.84EE762 pKa = 3.35VFAQVKK768 pKa = 9.02QMYY771 pKa = 7.48KK772 pKa = 9.69TPTLKK777 pKa = 10.84YY778 pKa = 9.33FGGFNFSQILPDD790 pKa = 3.84PLKK793 pKa = 8.1PTKK796 pKa = 10.2RR797 pKa = 11.84SFIEE801 pKa = 4.3DD802 pKa = 3.61LLFNKK807 pKa = 7.99VTLADD812 pKa = 3.57AGFMKK817 pKa = 10.37QYY819 pKa = 10.74GEE821 pKa = 4.31CLGDD825 pKa = 3.25INARR829 pKa = 11.84DD830 pKa = 4.56LICAQKK836 pKa = 10.77FNGLTVLPPLLTDD849 pKa = 4.9DD850 pKa = 4.88MIAAYY855 pKa = 8.09TAALVSGTATAGWTFGAGAALQIPFAMQMAYY886 pKa = 10.08RR887 pKa = 11.84FNGIGVTQNVLYY899 pKa = 10.46EE900 pKa = 4.09NQKK903 pKa = 10.62QIANQFNKK911 pKa = 10.46AISQIQEE918 pKa = 4.19SLTTTSTALGKK929 pKa = 10.37LQDD932 pKa = 3.95VVNQNAQALNTLVKK946 pKa = 10.38QLSSNFGAISSVLNDD961 pKa = 3.17ILSRR965 pKa = 11.84LDD967 pKa = 3.31KK968 pKa = 11.45VEE970 pKa = 5.19AEE972 pKa = 4.28VQIDD976 pKa = 3.56RR977 pKa = 11.84LITGRR982 pKa = 11.84LQSLQTYY989 pKa = 7.4VTQQLIRR996 pKa = 11.84AAEE999 pKa = 3.85IRR1001 pKa = 11.84ASANLAATKK1010 pKa = 9.96MSEE1013 pKa = 4.28CVLGQSKK1020 pKa = 10.12RR1021 pKa = 11.84VDD1023 pKa = 4.0FCGKK1027 pKa = 9.92GYY1029 pKa = 10.57HH1030 pKa = 6.84LMSFPQAAPHH1040 pKa = 5.76GVVFLHH1046 pKa = 5.25VTYY1049 pKa = 10.82VPSQEE1054 pKa = 4.9RR1055 pKa = 11.84NFTTAPAICHH1065 pKa = 5.73EE1066 pKa = 4.45GKK1068 pKa = 10.56AYY1070 pKa = 9.93FPRR1073 pKa = 11.84EE1074 pKa = 3.72GVFVFNGTSWFITQRR1089 pKa = 11.84NFFSPQIITTDD1100 pKa = 3.16NTFVSGNCDD1109 pKa = 3.2VVIGIINNTVYY1120 pKa = 10.97DD1121 pKa = 4.22PLQPEE1126 pKa = 4.19LDD1128 pKa = 3.85SFKK1131 pKa = 11.4EE1132 pKa = 3.91EE1133 pKa = 3.56LDD1135 pKa = 3.63KK1136 pKa = 11.53YY1137 pKa = 10.15FKK1139 pKa = 10.99NHH1141 pKa = 6.01TSPDD1145 pKa = 3.14VDD1147 pKa = 4.4LGDD1150 pKa = 3.31ISGINASVVNIQKK1163 pKa = 10.63EE1164 pKa = 3.92IDD1166 pKa = 3.67RR1167 pKa = 11.84LNEE1170 pKa = 3.5VAKK1173 pKa = 10.61NLNEE1177 pKa = 4.14SLIDD1181 pKa = 3.75LQEE1184 pKa = 4.17LGKK1187 pKa = 10.54YY1188 pKa = 6.98EE1189 pKa = 5.3QYY1191 pKa = 10.78IKK1193 pKa = 10.03WPWYY1197 pKa = 8.39VWLGFIAGLIAIVMVTILLCCMTSCCSCLKK1227 pKa = 10.16GACSCGSCCKK1237 pKa = 10.16FDD1239 pKa = 4.43EE1240 pKa = 5.39DD1241 pKa = 5.38DD1242 pKa = 4.38SEE1244 pKa = 4.36PVLKK1248 pKa = 10.36GVKK1251 pKa = 9.35LHH1253 pKa = 5.3YY1254 pKa = 9.44TT1255 pKa = 3.44
Molecular weight: 139.13 kDa
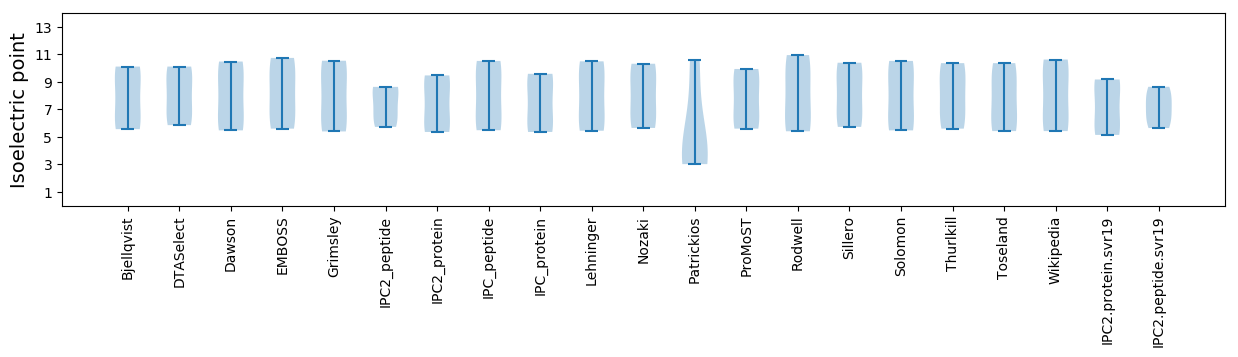
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6UZF3|Q6UZF3_SARS Membrane protein OS=SARS coronavirus PUMC02 OX=253434 GN=M PE=3 SV=1
MM1 pKa = 7.81SDD3 pKa = 4.42NGPQSNQRR11 pKa = 11.84SAPRR15 pKa = 11.84ITFGGPTDD23 pKa = 3.81STDD26 pKa = 3.04NNQNGGRR33 pKa = 11.84NGARR37 pKa = 11.84PKK39 pKa = 10.13QRR41 pKa = 11.84RR42 pKa = 11.84PQGLPNNTASWFTALTQHH60 pKa = 6.06GKK62 pKa = 10.36EE63 pKa = 3.96EE64 pKa = 4.07LRR66 pKa = 11.84FPRR69 pKa = 11.84GQGVPINTNSGPDD82 pKa = 3.62DD83 pKa = 3.51QIGYY87 pKa = 8.13YY88 pKa = 9.97RR89 pKa = 11.84RR90 pKa = 11.84ATRR93 pKa = 11.84RR94 pKa = 11.84VRR96 pKa = 11.84GGDD99 pKa = 3.09GKK101 pKa = 10.39MKK103 pKa = 9.92EE104 pKa = 4.5LSPRR108 pKa = 11.84WYY110 pKa = 10.01FYY112 pKa = 11.5YY113 pKa = 10.73LGTGPEE119 pKa = 3.63ASLPYY124 pKa = 9.9GANKK128 pKa = 9.97EE129 pKa = 4.24GIVWVATEE137 pKa = 4.37GALNTPKK144 pKa = 10.91DD145 pKa = 4.0HH146 pKa = 7.46IGTRR150 pKa = 11.84NPNNNAATVLQLPQGTTLPKK170 pKa = 10.34GFYY173 pKa = 10.67AEE175 pKa = 4.5GSRR178 pKa = 11.84GGSQASSRR186 pKa = 11.84SSSRR190 pKa = 11.84SRR192 pKa = 11.84GNSRR196 pKa = 11.84NSTPGSSRR204 pKa = 11.84GNSPARR210 pKa = 11.84MASGGGEE217 pKa = 3.84TALALLLLDD226 pKa = 4.89RR227 pKa = 11.84LNQLEE232 pKa = 4.58SKK234 pKa = 10.86VSGKK238 pKa = 10.04GQQQQGQTVTKK249 pKa = 9.98KK250 pKa = 10.44SAAEE254 pKa = 3.73ASKK257 pKa = 10.76KK258 pKa = 9.61PRR260 pKa = 11.84QKK262 pKa = 9.66RR263 pKa = 11.84TATKK267 pKa = 10.12QYY269 pKa = 11.12NVTQAFGRR277 pKa = 11.84RR278 pKa = 11.84GPEE281 pKa = 3.4QTQGNFGDD289 pKa = 3.74QDD291 pKa = 5.15LIRR294 pKa = 11.84QGTDD298 pKa = 2.96YY299 pKa = 11.27KK300 pKa = 10.5HH301 pKa = 6.32WPQIAQFAPSASAFFGMSRR320 pKa = 11.84IGMEE324 pKa = 4.16VTPSGTWLTYY334 pKa = 10.42HH335 pKa = 7.06GAIKK339 pKa = 10.61LDD341 pKa = 4.18DD342 pKa = 4.89KK343 pKa = 11.64DD344 pKa = 3.91PQFKK348 pKa = 11.14DD349 pKa = 3.7NVILLNKK356 pKa = 10.01HH357 pKa = 4.67IDD359 pKa = 3.35AYY361 pKa = 9.77KK362 pKa = 9.88TFPPTEE368 pKa = 3.95PKK370 pKa = 9.88KK371 pKa = 10.6DD372 pKa = 3.48KK373 pKa = 10.73KK374 pKa = 10.99KK375 pKa = 9.88KK376 pKa = 8.29TDD378 pKa = 3.29EE379 pKa = 4.34AQPLPQRR386 pKa = 11.84QKK388 pKa = 10.35KK389 pKa = 8.88QPTVTLLPAADD400 pKa = 3.84MDD402 pKa = 4.26DD403 pKa = 5.3FSRR406 pKa = 11.84QLQNSMSGASADD418 pKa = 3.63STQAA422 pKa = 3.04
MM1 pKa = 7.81SDD3 pKa = 4.42NGPQSNQRR11 pKa = 11.84SAPRR15 pKa = 11.84ITFGGPTDD23 pKa = 3.81STDD26 pKa = 3.04NNQNGGRR33 pKa = 11.84NGARR37 pKa = 11.84PKK39 pKa = 10.13QRR41 pKa = 11.84RR42 pKa = 11.84PQGLPNNTASWFTALTQHH60 pKa = 6.06GKK62 pKa = 10.36EE63 pKa = 3.96EE64 pKa = 4.07LRR66 pKa = 11.84FPRR69 pKa = 11.84GQGVPINTNSGPDD82 pKa = 3.62DD83 pKa = 3.51QIGYY87 pKa = 8.13YY88 pKa = 9.97RR89 pKa = 11.84RR90 pKa = 11.84ATRR93 pKa = 11.84RR94 pKa = 11.84VRR96 pKa = 11.84GGDD99 pKa = 3.09GKK101 pKa = 10.39MKK103 pKa = 9.92EE104 pKa = 4.5LSPRR108 pKa = 11.84WYY110 pKa = 10.01FYY112 pKa = 11.5YY113 pKa = 10.73LGTGPEE119 pKa = 3.63ASLPYY124 pKa = 9.9GANKK128 pKa = 9.97EE129 pKa = 4.24GIVWVATEE137 pKa = 4.37GALNTPKK144 pKa = 10.91DD145 pKa = 4.0HH146 pKa = 7.46IGTRR150 pKa = 11.84NPNNNAATVLQLPQGTTLPKK170 pKa = 10.34GFYY173 pKa = 10.67AEE175 pKa = 4.5GSRR178 pKa = 11.84GGSQASSRR186 pKa = 11.84SSSRR190 pKa = 11.84SRR192 pKa = 11.84GNSRR196 pKa = 11.84NSTPGSSRR204 pKa = 11.84GNSPARR210 pKa = 11.84MASGGGEE217 pKa = 3.84TALALLLLDD226 pKa = 4.89RR227 pKa = 11.84LNQLEE232 pKa = 4.58SKK234 pKa = 10.86VSGKK238 pKa = 10.04GQQQQGQTVTKK249 pKa = 9.98KK250 pKa = 10.44SAAEE254 pKa = 3.73ASKK257 pKa = 10.76KK258 pKa = 9.61PRR260 pKa = 11.84QKK262 pKa = 9.66RR263 pKa = 11.84TATKK267 pKa = 10.12QYY269 pKa = 11.12NVTQAFGRR277 pKa = 11.84RR278 pKa = 11.84GPEE281 pKa = 3.4QTQGNFGDD289 pKa = 3.74QDD291 pKa = 5.15LIRR294 pKa = 11.84QGTDD298 pKa = 2.96YY299 pKa = 11.27KK300 pKa = 10.5HH301 pKa = 6.32WPQIAQFAPSASAFFGMSRR320 pKa = 11.84IGMEE324 pKa = 4.16VTPSGTWLTYY334 pKa = 10.42HH335 pKa = 7.06GAIKK339 pKa = 10.61LDD341 pKa = 4.18DD342 pKa = 4.89KK343 pKa = 11.64DD344 pKa = 3.91PQFKK348 pKa = 11.14DD349 pKa = 3.7NVILLNKK356 pKa = 10.01HH357 pKa = 4.67IDD359 pKa = 3.35AYY361 pKa = 9.77KK362 pKa = 9.88TFPPTEE368 pKa = 3.95PKK370 pKa = 9.88KK371 pKa = 10.6DD372 pKa = 3.48KK373 pKa = 10.73KK374 pKa = 10.99KK375 pKa = 9.88KK376 pKa = 8.29TDD378 pKa = 3.29EE379 pKa = 4.34AQPLPQRR386 pKa = 11.84QKK388 pKa = 10.35KK389 pKa = 8.88QPTVTLLPAADD400 pKa = 3.84MDD402 pKa = 4.26DD403 pKa = 5.3FSRR406 pKa = 11.84QLQNSMSGASADD418 pKa = 3.63STQAA422 pKa = 3.04
Molecular weight: 46.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8971 |
221 |
7073 |
2242.8 |
250.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.223 ± 0.206 | 3.077 ± 0.488 |
5.54 ± 0.282 | 4.57 ± 0.647 |
4.871 ± 0.562 | 6.22 ± 0.646 |
2.04 ± 0.428 | 5.016 ± 0.519 |
5.685 ± 0.458 | 9.252 ± 0.692 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.352 ± 0.311 | 5.406 ± 0.458 |
4.091 ± 0.471 | 3.656 ± 0.703 |
3.835 ± 0.58 | 6.699 ± 0.45 |
7.134 ± 0.301 | 7.781 ± 0.849 |
1.115 ± 0.194 | 4.437 ± 0.298 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
