
Cycloclasticus sp. symbiont of Poecilosclerida sp. M
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Piscirickettsiaceae; Cycloclasticus; unclassified Cycloclasticus
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
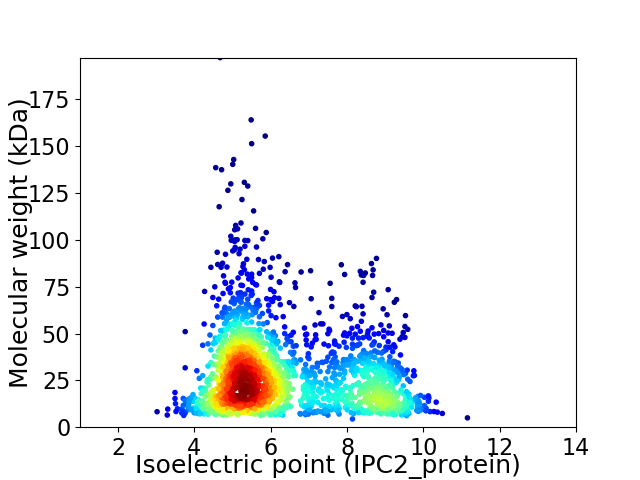
Virtual 2D-PAGE plot for 1993 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X1QEJ3|A0A1X1QEJ3_9GAMM Pimeloyl-[acyl-carrier protein] methyl ester esterase OS=Cycloclasticus sp. symbiont of Poecilosclerida sp. M OX=1840537 GN=bioH PE=3 SV=1
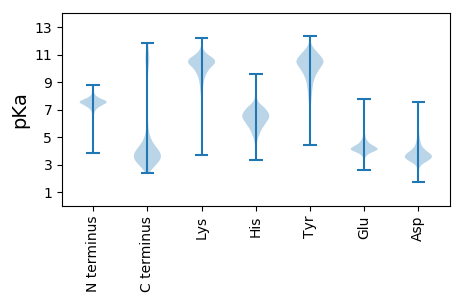
MM1 pKa = 6.58EE2 pKa = 5.86TIRR5 pKa = 11.84KK6 pKa = 6.96TPLSLGVAAGLGLLLASTGAMAISTGTDD34 pKa = 3.09LNLSAKK40 pKa = 9.9ARR42 pKa = 11.84AGGMAGAAYY51 pKa = 8.83TMPQEE56 pKa = 3.77ASAAVFGNPATLTQFKK72 pKa = 9.74GINMNFGASYY82 pKa = 10.79LGLKK86 pKa = 10.23SIDD89 pKa = 3.62IEE91 pKa = 4.43TSSTIAALGGGVSTSSKK108 pKa = 10.88SDD110 pKa = 3.04ADD112 pKa = 3.69NYY114 pKa = 10.15IVPDD118 pKa = 4.6FGLTLQVSPNLVLGTGLEE136 pKa = 4.06VDD138 pKa = 3.88AGLGADD144 pKa = 4.08YY145 pKa = 10.81RR146 pKa = 11.84DD147 pKa = 4.49DD148 pKa = 5.29PISLLGTGLVTIPLNVEE165 pKa = 4.53LISFNANLAAAYY177 pKa = 8.63QATEE181 pKa = 3.97KK182 pKa = 10.9LSIGGAVTIGFALAQLGTTGPTGGGAGLTALLGSDD217 pKa = 4.7APTDD221 pKa = 3.88LDD223 pKa = 4.41GPGPLPAIGANLGINDD239 pKa = 4.19FGGTTSSVHH248 pKa = 7.35DD249 pKa = 3.6IGFSGSLGATYY260 pKa = 9.76EE261 pKa = 4.16VQEE264 pKa = 4.21GVMLSAAYY272 pKa = 7.38KK273 pKa = 9.64TKK275 pKa = 10.67HH276 pKa = 5.95EE277 pKa = 4.48YY278 pKa = 10.98NFTNILYY285 pKa = 10.07QDD287 pKa = 3.51TSLNGPSGNGAGLGSGAFGPDD308 pKa = 3.4YY309 pKa = 10.92QDD311 pKa = 4.58LSVAQPAEE319 pKa = 4.37VIVGVAFDD327 pKa = 3.78NVIAQGLLVEE337 pKa = 5.39ADD339 pKa = 3.92VVWKK343 pKa = 10.12NWSDD347 pKa = 2.98AATYY351 pKa = 9.87EE352 pKa = 4.35NAYY355 pKa = 10.61DD356 pKa = 3.81DD357 pKa = 4.17QFLFLLGAQYY367 pKa = 7.91EE368 pKa = 4.44TGDD371 pKa = 2.86WSFRR375 pKa = 11.84AGYY378 pKa = 10.09SYY380 pKa = 11.68AEE382 pKa = 5.37DD383 pKa = 3.8ILRR386 pKa = 11.84SDD388 pKa = 4.64PDD390 pKa = 3.58TTLDD394 pKa = 3.66SLVGLGTLPVGDD406 pKa = 4.78GGPFGNDD413 pKa = 2.32IVKK416 pKa = 10.15IAQATLLPVIWNHH429 pKa = 6.65TITAGVGYY437 pKa = 10.09AITDD441 pKa = 3.56AVSIDD446 pKa = 3.6AYY448 pKa = 10.38AAYY451 pKa = 10.34AFGEE455 pKa = 4.57EE456 pKa = 4.33EE457 pKa = 4.37TVDD460 pKa = 3.64LPKK463 pKa = 10.57IQDD466 pKa = 3.25AAVAYY471 pKa = 7.07VVPVTEE477 pKa = 4.76LQLEE481 pKa = 4.21AQVDD485 pKa = 3.94YY486 pKa = 11.16EE487 pKa = 4.55FMVGAGINIALPP499 pKa = 3.76
MM1 pKa = 6.58EE2 pKa = 5.86TIRR5 pKa = 11.84KK6 pKa = 6.96TPLSLGVAAGLGLLLASTGAMAISTGTDD34 pKa = 3.09LNLSAKK40 pKa = 9.9ARR42 pKa = 11.84AGGMAGAAYY51 pKa = 8.83TMPQEE56 pKa = 3.77ASAAVFGNPATLTQFKK72 pKa = 9.74GINMNFGASYY82 pKa = 10.79LGLKK86 pKa = 10.23SIDD89 pKa = 3.62IEE91 pKa = 4.43TSSTIAALGGGVSTSSKK108 pKa = 10.88SDD110 pKa = 3.04ADD112 pKa = 3.69NYY114 pKa = 10.15IVPDD118 pKa = 4.6FGLTLQVSPNLVLGTGLEE136 pKa = 4.06VDD138 pKa = 3.88AGLGADD144 pKa = 4.08YY145 pKa = 10.81RR146 pKa = 11.84DD147 pKa = 4.49DD148 pKa = 5.29PISLLGTGLVTIPLNVEE165 pKa = 4.53LISFNANLAAAYY177 pKa = 8.63QATEE181 pKa = 3.97KK182 pKa = 10.9LSIGGAVTIGFALAQLGTTGPTGGGAGLTALLGSDD217 pKa = 4.7APTDD221 pKa = 3.88LDD223 pKa = 4.41GPGPLPAIGANLGINDD239 pKa = 4.19FGGTTSSVHH248 pKa = 7.35DD249 pKa = 3.6IGFSGSLGATYY260 pKa = 9.76EE261 pKa = 4.16VQEE264 pKa = 4.21GVMLSAAYY272 pKa = 7.38KK273 pKa = 9.64TKK275 pKa = 10.67HH276 pKa = 5.95EE277 pKa = 4.48YY278 pKa = 10.98NFTNILYY285 pKa = 10.07QDD287 pKa = 3.51TSLNGPSGNGAGLGSGAFGPDD308 pKa = 3.4YY309 pKa = 10.92QDD311 pKa = 4.58LSVAQPAEE319 pKa = 4.37VIVGVAFDD327 pKa = 3.78NVIAQGLLVEE337 pKa = 5.39ADD339 pKa = 3.92VVWKK343 pKa = 10.12NWSDD347 pKa = 2.98AATYY351 pKa = 9.87EE352 pKa = 4.35NAYY355 pKa = 10.61DD356 pKa = 3.81DD357 pKa = 4.17QFLFLLGAQYY367 pKa = 7.91EE368 pKa = 4.44TGDD371 pKa = 2.86WSFRR375 pKa = 11.84AGYY378 pKa = 10.09SYY380 pKa = 11.68AEE382 pKa = 5.37DD383 pKa = 3.8ILRR386 pKa = 11.84SDD388 pKa = 4.64PDD390 pKa = 3.58TTLDD394 pKa = 3.66SLVGLGTLPVGDD406 pKa = 4.78GGPFGNDD413 pKa = 2.32IVKK416 pKa = 10.15IAQATLLPVIWNHH429 pKa = 6.65TITAGVGYY437 pKa = 10.09AITDD441 pKa = 3.56AVSIDD446 pKa = 3.6AYY448 pKa = 10.38AAYY451 pKa = 10.34AFGEE455 pKa = 4.57EE456 pKa = 4.33EE457 pKa = 4.37TVDD460 pKa = 3.64LPKK463 pKa = 10.57IQDD466 pKa = 3.25AAVAYY471 pKa = 7.07VVPVTEE477 pKa = 4.76LQLEE481 pKa = 4.21AQVDD485 pKa = 3.94YY486 pKa = 11.16EE487 pKa = 4.55FMVGAGINIALPP499 pKa = 3.76
Molecular weight: 51.04 kDa
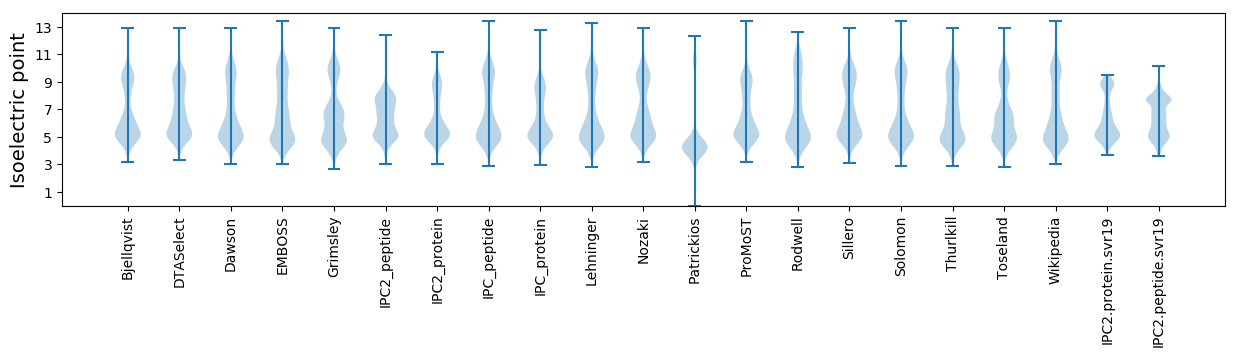
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X1QHJ0|A0A1X1QHJ0_9GAMM CopG family transcriptional regulator OS=Cycloclasticus sp. symbiont of Poecilosclerida sp. M OX=1840537 GN=A6F71_06825 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.01GRR39 pKa = 11.84AVLSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.01GRR39 pKa = 11.84AVLSAA44 pKa = 3.96
Molecular weight: 4.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
544741 |
35 |
1771 |
273.3 |
30.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.493 ± 0.054 | 1.227 ± 0.022 |
5.57 ± 0.04 | 6.398 ± 0.06 |
4.004 ± 0.04 | 7.131 ± 0.053 |
2.261 ± 0.027 | 6.576 ± 0.038 |
6.025 ± 0.052 | 10.276 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.577 ± 0.028 | 4.055 ± 0.037 |
3.915 ± 0.03 | 4.001 ± 0.039 |
4.62 ± 0.044 | 6.561 ± 0.042 |
5.365 ± 0.038 | 7.06 ± 0.049 |
1.116 ± 0.021 | 2.77 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
