
Schaedlerella arabinosiphila
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Schaedlerella
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
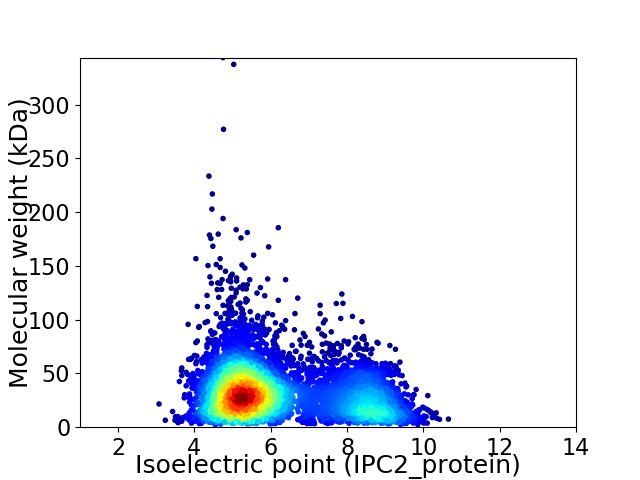
Virtual 2D-PAGE plot for 5980 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|N2A3L7|N2A3L7_9FIRM Ethanolamine utilization protein EutN OS=Schaedlerella arabinosiphila OX=2044587 GN=C824_05627 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.51KK3 pKa = 10.05FVALFLALTLTCGLAACGGGGNSGGGDD30 pKa = 3.74SNDD33 pKa = 3.71TGGSGASTEE42 pKa = 4.07EE43 pKa = 4.17STDD46 pKa = 3.29GDD48 pKa = 3.89EE49 pKa = 5.73KK50 pKa = 11.07EE51 pKa = 4.76DD52 pKa = 3.58SGSGEE57 pKa = 3.69KK58 pKa = 10.46VAINVIAAQYY68 pKa = 8.87GQNTTKK74 pKa = 9.08WWSDD78 pKa = 3.22FVKK81 pKa = 10.58DD82 pKa = 4.04FTTEE86 pKa = 3.87YY87 pKa = 10.42PNIDD91 pKa = 3.3LTVEE95 pKa = 3.93VEE97 pKa = 4.12SWNDD101 pKa = 2.46IYY103 pKa = 11.3TVVNTRR109 pKa = 11.84ISNNDD114 pKa = 3.06APDD117 pKa = 3.41ILNIDD122 pKa = 3.48VFANYY127 pKa = 9.66QADD130 pKa = 4.2DD131 pKa = 4.26LLLPAEE137 pKa = 4.98DD138 pKa = 3.96FVSDD142 pKa = 3.4EE143 pKa = 4.92TYY145 pKa = 11.32KK146 pKa = 11.09KK147 pKa = 9.54MYY149 pKa = 9.48PAFLEE154 pKa = 3.91QSNVDD159 pKa = 3.41GTIWAIPDD167 pKa = 3.61LASARR172 pKa = 11.84ALYY175 pKa = 10.02VNKK178 pKa = 10.43DD179 pKa = 3.03ILDD182 pKa = 3.93AAKK185 pKa = 10.86VEE187 pKa = 5.25IPTTWDD193 pKa = 3.31EE194 pKa = 4.22LKK196 pKa = 10.56KK197 pKa = 10.76ACEE200 pKa = 4.18AIKK203 pKa = 10.5AYY205 pKa = 10.15DD206 pKa = 3.53SKK208 pKa = 10.89IYY210 pKa = 9.79PWGIDD215 pKa = 3.14MTTDD219 pKa = 2.85EE220 pKa = 4.57GQAAFAYY227 pKa = 7.2YY228 pKa = 8.91TWNNGGGFVDD238 pKa = 5.26EE239 pKa = 4.97DD240 pKa = 4.21NNWALNSDD248 pKa = 4.09EE249 pKa = 4.56NVEE252 pKa = 4.03AVEE255 pKa = 4.26FAISLVKK262 pKa = 10.53DD263 pKa = 4.56GYY265 pKa = 9.47TNSDD269 pKa = 3.39PSNDD273 pKa = 3.13TRR275 pKa = 11.84YY276 pKa = 10.71DD277 pKa = 3.61LQDD280 pKa = 3.21MFGAGQVAMMIGPNSIPTYY299 pKa = 9.99IADD302 pKa = 3.56GGYY305 pKa = 9.63SVNYY309 pKa = 8.79EE310 pKa = 4.03VAPIPTNGDD319 pKa = 3.48NEE321 pKa = 4.65SVSAGVMDD329 pKa = 5.53RR330 pKa = 11.84FMCFDD335 pKa = 3.57NGYY338 pKa = 10.62SDD340 pKa = 5.76AEE342 pKa = 4.16LEE344 pKa = 4.39AIKK347 pKa = 9.97TFFDD351 pKa = 3.63YY352 pKa = 10.25FYY354 pKa = 10.87EE355 pKa = 4.14DD356 pKa = 2.68TRR358 pKa = 11.84YY359 pKa = 10.59SDD361 pKa = 3.26WVLMEE366 pKa = 4.43GFLPATSTGGEE377 pKa = 3.91ILAEE381 pKa = 4.46ADD383 pKa = 3.07EE384 pKa = 5.56SMASWVDD391 pKa = 3.24IVGSCNFYY399 pKa = 9.29PTAKK403 pKa = 10.06TEE405 pKa = 3.99WDD407 pKa = 3.59DD408 pKa = 3.73VKK410 pKa = 11.15QGVINVEE417 pKa = 3.95QQALLDD423 pKa = 4.22GNVKK427 pKa = 10.19EE428 pKa = 5.75LLDD431 pKa = 3.83EE432 pKa = 4.48LQAEE436 pKa = 4.41IAEE439 pKa = 4.32
MM1 pKa = 7.5KK2 pKa = 10.51KK3 pKa = 10.05FVALFLALTLTCGLAACGGGGNSGGGDD30 pKa = 3.74SNDD33 pKa = 3.71TGGSGASTEE42 pKa = 4.07EE43 pKa = 4.17STDD46 pKa = 3.29GDD48 pKa = 3.89EE49 pKa = 5.73KK50 pKa = 11.07EE51 pKa = 4.76DD52 pKa = 3.58SGSGEE57 pKa = 3.69KK58 pKa = 10.46VAINVIAAQYY68 pKa = 8.87GQNTTKK74 pKa = 9.08WWSDD78 pKa = 3.22FVKK81 pKa = 10.58DD82 pKa = 4.04FTTEE86 pKa = 3.87YY87 pKa = 10.42PNIDD91 pKa = 3.3LTVEE95 pKa = 3.93VEE97 pKa = 4.12SWNDD101 pKa = 2.46IYY103 pKa = 11.3TVVNTRR109 pKa = 11.84ISNNDD114 pKa = 3.06APDD117 pKa = 3.41ILNIDD122 pKa = 3.48VFANYY127 pKa = 9.66QADD130 pKa = 4.2DD131 pKa = 4.26LLLPAEE137 pKa = 4.98DD138 pKa = 3.96FVSDD142 pKa = 3.4EE143 pKa = 4.92TYY145 pKa = 11.32KK146 pKa = 11.09KK147 pKa = 9.54MYY149 pKa = 9.48PAFLEE154 pKa = 3.91QSNVDD159 pKa = 3.41GTIWAIPDD167 pKa = 3.61LASARR172 pKa = 11.84ALYY175 pKa = 10.02VNKK178 pKa = 10.43DD179 pKa = 3.03ILDD182 pKa = 3.93AAKK185 pKa = 10.86VEE187 pKa = 5.25IPTTWDD193 pKa = 3.31EE194 pKa = 4.22LKK196 pKa = 10.56KK197 pKa = 10.76ACEE200 pKa = 4.18AIKK203 pKa = 10.5AYY205 pKa = 10.15DD206 pKa = 3.53SKK208 pKa = 10.89IYY210 pKa = 9.79PWGIDD215 pKa = 3.14MTTDD219 pKa = 2.85EE220 pKa = 4.57GQAAFAYY227 pKa = 7.2YY228 pKa = 8.91TWNNGGGFVDD238 pKa = 5.26EE239 pKa = 4.97DD240 pKa = 4.21NNWALNSDD248 pKa = 4.09EE249 pKa = 4.56NVEE252 pKa = 4.03AVEE255 pKa = 4.26FAISLVKK262 pKa = 10.53DD263 pKa = 4.56GYY265 pKa = 9.47TNSDD269 pKa = 3.39PSNDD273 pKa = 3.13TRR275 pKa = 11.84YY276 pKa = 10.71DD277 pKa = 3.61LQDD280 pKa = 3.21MFGAGQVAMMIGPNSIPTYY299 pKa = 9.99IADD302 pKa = 3.56GGYY305 pKa = 9.63SVNYY309 pKa = 8.79EE310 pKa = 4.03VAPIPTNGDD319 pKa = 3.48NEE321 pKa = 4.65SVSAGVMDD329 pKa = 5.53RR330 pKa = 11.84FMCFDD335 pKa = 3.57NGYY338 pKa = 10.62SDD340 pKa = 5.76AEE342 pKa = 4.16LEE344 pKa = 4.39AIKK347 pKa = 9.97TFFDD351 pKa = 3.63YY352 pKa = 10.25FYY354 pKa = 10.87EE355 pKa = 4.14DD356 pKa = 2.68TRR358 pKa = 11.84YY359 pKa = 10.59SDD361 pKa = 3.26WVLMEE366 pKa = 4.43GFLPATSTGGEE377 pKa = 3.91ILAEE381 pKa = 4.46ADD383 pKa = 3.07EE384 pKa = 5.56SMASWVDD391 pKa = 3.24IVGSCNFYY399 pKa = 9.29PTAKK403 pKa = 10.06TEE405 pKa = 3.99WDD407 pKa = 3.59DD408 pKa = 3.73VKK410 pKa = 11.15QGVINVEE417 pKa = 3.95QQALLDD423 pKa = 4.22GNVKK427 pKa = 10.19EE428 pKa = 5.75LLDD431 pKa = 3.83EE432 pKa = 4.48LQAEE436 pKa = 4.41IAEE439 pKa = 4.32
Molecular weight: 48.07 kDa
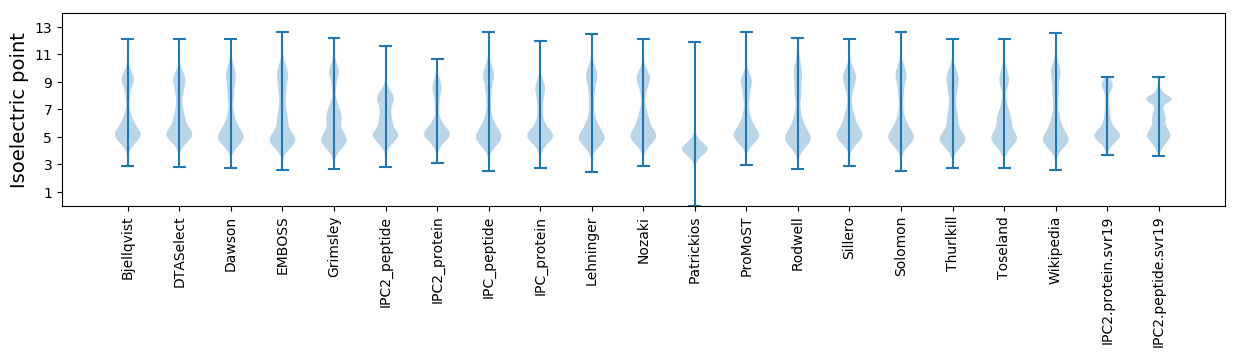
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|N1ZVG7|N1ZVG7_9FIRM HTH cro/C1-type domain-containing protein OS=Schaedlerella arabinosiphila OX=2044587 GN=C824_05465 PE=4 SV=1
MM1 pKa = 7.17FVCNNRR7 pKa = 11.84GLSFGIAFYY16 pKa = 10.47LYY18 pKa = 9.6HH19 pKa = 6.79FCLIHH24 pKa = 6.89SDD26 pKa = 3.3VRR28 pKa = 11.84ISRR31 pKa = 11.84RR32 pKa = 11.84IGKK35 pKa = 8.94SRR37 pKa = 11.84NGVLILMWLWIRR49 pKa = 11.84FPLLKK54 pKa = 10.51AVFQGLRR61 pKa = 11.84AYY63 pKa = 10.25LRR65 pKa = 11.84QYY67 pKa = 9.37QTDD70 pKa = 3.34QDD72 pKa = 3.39KK73 pKa = 11.21KK74 pKa = 10.76MRR76 pKa = 11.84DD77 pKa = 2.97RR78 pKa = 11.84WTDD81 pKa = 2.65II82 pKa = 3.42
MM1 pKa = 7.17FVCNNRR7 pKa = 11.84GLSFGIAFYY16 pKa = 10.47LYY18 pKa = 9.6HH19 pKa = 6.79FCLIHH24 pKa = 6.89SDD26 pKa = 3.3VRR28 pKa = 11.84ISRR31 pKa = 11.84RR32 pKa = 11.84IGKK35 pKa = 8.94SRR37 pKa = 11.84NGVLILMWLWIRR49 pKa = 11.84FPLLKK54 pKa = 10.51AVFQGLRR61 pKa = 11.84AYY63 pKa = 10.25LRR65 pKa = 11.84QYY67 pKa = 9.37QTDD70 pKa = 3.34QDD72 pKa = 3.39KK73 pKa = 11.21KK74 pKa = 10.76MRR76 pKa = 11.84DD77 pKa = 2.97RR78 pKa = 11.84WTDD81 pKa = 2.65II82 pKa = 3.42
Molecular weight: 9.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1817401 |
21 |
3066 |
303.9 |
34.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.538 ± 0.04 | 1.553 ± 0.014 |
5.486 ± 0.025 | 8.026 ± 0.037 |
4.165 ± 0.025 | 7.152 ± 0.038 |
1.804 ± 0.014 | 6.978 ± 0.034 |
6.477 ± 0.031 | 9.15 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.126 ± 0.018 | 4.089 ± 0.022 |
3.37 ± 0.017 | 3.454 ± 0.017 |
5.127 ± 0.029 | 5.818 ± 0.032 |
5.063 ± 0.022 | 6.429 ± 0.026 |
0.998 ± 0.012 | 4.197 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
