
Lewinella sp. 4G2
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Saprospiria; Saprospirales; Lewinellaceae; Lewinella; unclassified Lewinella
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
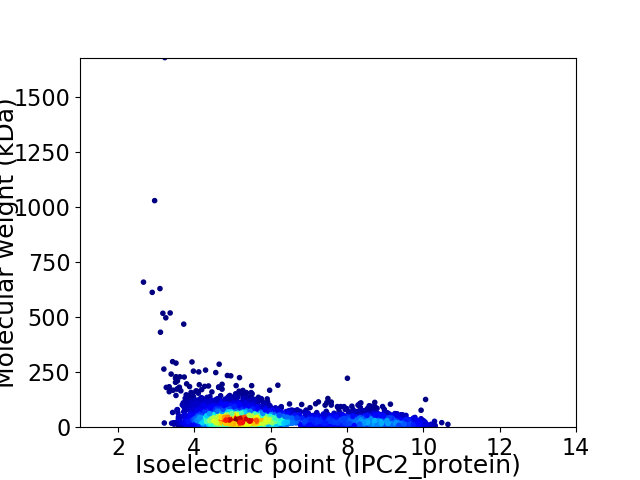
Virtual 2D-PAGE plot for 3730 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A180ESP5|A0A180ESP5_9BACT 50S ribosomal protein L18 OS=Lewinella sp. 4G2 OX=1803372 GN=rplR PE=3 SV=1
MM1 pKa = 7.5TFFKK5 pKa = 10.84KK6 pKa = 10.44SITLICLVLLASTLSGQSFSDD27 pKa = 4.21DD28 pKa = 3.34FAADD32 pKa = 4.15LSNWPVTAGWQWDD45 pKa = 3.7ADD47 pKa = 3.74GKK49 pKa = 11.04ADD51 pKa = 5.58NIPNVNWNNRR61 pKa = 11.84SRR63 pKa = 11.84LRR65 pKa = 11.84SASEE69 pKa = 3.95GGAALYY75 pKa = 10.47DD76 pKa = 3.79SSTGAGRR83 pKa = 11.84LTSRR87 pKa = 11.84IFTPDD92 pKa = 3.34TEE94 pKa = 4.86PGLFVSFYY102 pKa = 10.72HH103 pKa = 6.23YY104 pKa = 10.23FQNQGGQIRR113 pKa = 11.84LQVIEE118 pKa = 4.14SGGNVLYY125 pKa = 10.56DD126 pKa = 3.7EE127 pKa = 4.88PLVPNVVSDD136 pKa = 4.11EE137 pKa = 4.14EE138 pKa = 4.38TSAGNFQIIDD148 pKa = 3.69LSFLKK153 pKa = 10.37TNGLNDD159 pKa = 3.51FQISFEE165 pKa = 4.09VATNIEE171 pKa = 4.39FWLLDD176 pKa = 3.86DD177 pKa = 4.1VTVGTDD183 pKa = 3.18RR184 pKa = 11.84PAWPTFPIYY193 pKa = 10.62FGQQLTNFGIPFNVDD208 pKa = 2.87SLAMPFVPFEE218 pKa = 4.19VVLDD222 pKa = 4.83FAATATPAIRR232 pKa = 11.84DD233 pKa = 4.0AIRR236 pKa = 11.84AEE238 pKa = 3.8FDD240 pKa = 3.17IVSVEE245 pKa = 4.0SCVCDD250 pKa = 3.63RR251 pKa = 11.84LEE253 pKa = 3.71VWTMGGGVFFDD264 pKa = 4.21EE265 pKa = 5.1ATGQPLGPPGDD276 pKa = 3.37ILEE279 pKa = 4.49RR280 pKa = 11.84VLGSSTNSGAEE291 pKa = 3.79SGEE294 pKa = 3.77VDD296 pKa = 3.84NIEE299 pKa = 4.84LNVLTYY305 pKa = 10.99DD306 pKa = 3.98EE307 pKa = 5.42LLSNPNAPTSPLQSGDD323 pKa = 3.53LAFLTDD329 pKa = 3.96APAGAVKK336 pKa = 10.5VAVLDD341 pKa = 3.61TGLDD345 pKa = 3.58LDD347 pKa = 4.8HH348 pKa = 7.31PTLQPYY354 pKa = 10.09LYY356 pKa = 10.87KK357 pKa = 10.62NDD359 pKa = 4.36DD360 pKa = 3.84PLGDD364 pKa = 4.78GEE366 pKa = 6.07DD367 pKa = 4.63DD368 pKa = 5.41DD369 pKa = 6.38SNCQTDD375 pKa = 6.12DD376 pKa = 4.38LIGWNFVDD384 pKa = 5.45GNNNPDD390 pKa = 4.12DD391 pKa = 3.96GHH393 pKa = 7.07GHH395 pKa = 5.23GTHH398 pKa = 5.77VAGIVAQTVSEE409 pKa = 4.32CDD411 pKa = 3.09NCTVQIMPYY420 pKa = 8.33KK421 pKa = 8.54THH423 pKa = 7.6DD424 pKa = 3.51NFGVGTVFNAACAVLQASVYY444 pKa = 11.05DD445 pKa = 3.84EE446 pKa = 4.86ADD448 pKa = 3.51VINASWGFYY457 pKa = 10.18GQGSDD462 pKa = 2.76ILRR465 pKa = 11.84RR466 pKa = 11.84AIDD469 pKa = 3.26TAAVYY474 pKa = 9.8GALVVAAVGNDD485 pKa = 3.74SLNMVADD492 pKa = 4.08DD493 pKa = 4.54QFPATYY499 pKa = 9.68TSNTVMSVAAFDD511 pKa = 3.99FDD513 pKa = 4.34GVSEE517 pKa = 4.74LSLAPYY523 pKa = 10.64SNYY526 pKa = 9.97NPSFADD532 pKa = 2.9ISYY535 pKa = 10.29YY536 pKa = 9.49GTAINSALPGGGLGNKK552 pKa = 8.6TGTSMATPALSATAALHH569 pKa = 6.61LCMTPSLSAAALRR582 pKa = 11.84TEE584 pKa = 4.35ILNNAYY590 pKa = 10.37DD591 pKa = 3.68NSAEE595 pKa = 3.99LGTFIFNGRR604 pKa = 11.84IPDD607 pKa = 3.75IEE609 pKa = 4.81DD610 pKa = 3.29VCEE613 pKa = 4.02MTDD616 pKa = 3.34PGNDD620 pKa = 3.28GTAAPYY626 pKa = 9.45SACICDD632 pKa = 4.35DD633 pKa = 3.76EE634 pKa = 4.76QPSVVSVTADD644 pKa = 3.37SPEE647 pKa = 3.9GEE649 pKa = 4.08YY650 pKa = 10.27TIRR653 pKa = 11.84VLTLSGEE660 pKa = 4.14EE661 pKa = 4.09LNRR664 pKa = 11.84VDD666 pKa = 4.98NAPLEE671 pKa = 4.54AGQPLNVEE679 pKa = 4.8LGTDD683 pKa = 3.48EE684 pKa = 5.01PGTYY688 pKa = 10.38LIVIKK693 pKa = 10.73NDD695 pKa = 2.82LEE697 pKa = 4.38TRR699 pKa = 11.84VLRR702 pKa = 11.84VVRR705 pKa = 11.84QDD707 pKa = 2.85
MM1 pKa = 7.5TFFKK5 pKa = 10.84KK6 pKa = 10.44SITLICLVLLASTLSGQSFSDD27 pKa = 4.21DD28 pKa = 3.34FAADD32 pKa = 4.15LSNWPVTAGWQWDD45 pKa = 3.7ADD47 pKa = 3.74GKK49 pKa = 11.04ADD51 pKa = 5.58NIPNVNWNNRR61 pKa = 11.84SRR63 pKa = 11.84LRR65 pKa = 11.84SASEE69 pKa = 3.95GGAALYY75 pKa = 10.47DD76 pKa = 3.79SSTGAGRR83 pKa = 11.84LTSRR87 pKa = 11.84IFTPDD92 pKa = 3.34TEE94 pKa = 4.86PGLFVSFYY102 pKa = 10.72HH103 pKa = 6.23YY104 pKa = 10.23FQNQGGQIRR113 pKa = 11.84LQVIEE118 pKa = 4.14SGGNVLYY125 pKa = 10.56DD126 pKa = 3.7EE127 pKa = 4.88PLVPNVVSDD136 pKa = 4.11EE137 pKa = 4.14EE138 pKa = 4.38TSAGNFQIIDD148 pKa = 3.69LSFLKK153 pKa = 10.37TNGLNDD159 pKa = 3.51FQISFEE165 pKa = 4.09VATNIEE171 pKa = 4.39FWLLDD176 pKa = 3.86DD177 pKa = 4.1VTVGTDD183 pKa = 3.18RR184 pKa = 11.84PAWPTFPIYY193 pKa = 10.62FGQQLTNFGIPFNVDD208 pKa = 2.87SLAMPFVPFEE218 pKa = 4.19VVLDD222 pKa = 4.83FAATATPAIRR232 pKa = 11.84DD233 pKa = 4.0AIRR236 pKa = 11.84AEE238 pKa = 3.8FDD240 pKa = 3.17IVSVEE245 pKa = 4.0SCVCDD250 pKa = 3.63RR251 pKa = 11.84LEE253 pKa = 3.71VWTMGGGVFFDD264 pKa = 4.21EE265 pKa = 5.1ATGQPLGPPGDD276 pKa = 3.37ILEE279 pKa = 4.49RR280 pKa = 11.84VLGSSTNSGAEE291 pKa = 3.79SGEE294 pKa = 3.77VDD296 pKa = 3.84NIEE299 pKa = 4.84LNVLTYY305 pKa = 10.99DD306 pKa = 3.98EE307 pKa = 5.42LLSNPNAPTSPLQSGDD323 pKa = 3.53LAFLTDD329 pKa = 3.96APAGAVKK336 pKa = 10.5VAVLDD341 pKa = 3.61TGLDD345 pKa = 3.58LDD347 pKa = 4.8HH348 pKa = 7.31PTLQPYY354 pKa = 10.09LYY356 pKa = 10.87KK357 pKa = 10.62NDD359 pKa = 4.36DD360 pKa = 3.84PLGDD364 pKa = 4.78GEE366 pKa = 6.07DD367 pKa = 4.63DD368 pKa = 5.41DD369 pKa = 6.38SNCQTDD375 pKa = 6.12DD376 pKa = 4.38LIGWNFVDD384 pKa = 5.45GNNNPDD390 pKa = 4.12DD391 pKa = 3.96GHH393 pKa = 7.07GHH395 pKa = 5.23GTHH398 pKa = 5.77VAGIVAQTVSEE409 pKa = 4.32CDD411 pKa = 3.09NCTVQIMPYY420 pKa = 8.33KK421 pKa = 8.54THH423 pKa = 7.6DD424 pKa = 3.51NFGVGTVFNAACAVLQASVYY444 pKa = 11.05DD445 pKa = 3.84EE446 pKa = 4.86ADD448 pKa = 3.51VINASWGFYY457 pKa = 10.18GQGSDD462 pKa = 2.76ILRR465 pKa = 11.84RR466 pKa = 11.84AIDD469 pKa = 3.26TAAVYY474 pKa = 9.8GALVVAAVGNDD485 pKa = 3.74SLNMVADD492 pKa = 4.08DD493 pKa = 4.54QFPATYY499 pKa = 9.68TSNTVMSVAAFDD511 pKa = 3.99FDD513 pKa = 4.34GVSEE517 pKa = 4.74LSLAPYY523 pKa = 10.64SNYY526 pKa = 9.97NPSFADD532 pKa = 2.9ISYY535 pKa = 10.29YY536 pKa = 9.49GTAINSALPGGGLGNKK552 pKa = 8.6TGTSMATPALSATAALHH569 pKa = 6.61LCMTPSLSAAALRR582 pKa = 11.84TEE584 pKa = 4.35ILNNAYY590 pKa = 10.37DD591 pKa = 3.68NSAEE595 pKa = 3.99LGTFIFNGRR604 pKa = 11.84IPDD607 pKa = 3.75IEE609 pKa = 4.81DD610 pKa = 3.29VCEE613 pKa = 4.02MTDD616 pKa = 3.34PGNDD620 pKa = 3.28GTAAPYY626 pKa = 9.45SACICDD632 pKa = 4.35DD633 pKa = 3.76EE634 pKa = 4.76QPSVVSVTADD644 pKa = 3.37SPEE647 pKa = 3.9GEE649 pKa = 4.08YY650 pKa = 10.27TIRR653 pKa = 11.84VLTLSGEE660 pKa = 4.14EE661 pKa = 4.09LNRR664 pKa = 11.84VDD666 pKa = 4.98NAPLEE671 pKa = 4.54AGQPLNVEE679 pKa = 4.8LGTDD683 pKa = 3.48EE684 pKa = 5.01PGTYY688 pKa = 10.38LIVIKK693 pKa = 10.73NDD695 pKa = 2.82LEE697 pKa = 4.38TRR699 pKa = 11.84VLRR702 pKa = 11.84VVRR705 pKa = 11.84QDD707 pKa = 2.85
Molecular weight: 75.63 kDa
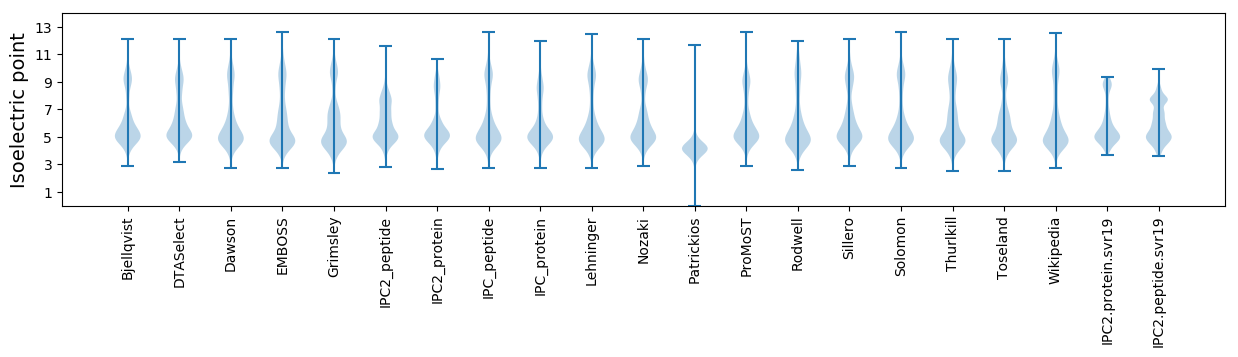
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A180EMX3|A0A180EMX3_9BACT Uncharacterized protein OS=Lewinella sp. 4G2 OX=1803372 GN=A3850_015780 PE=4 SV=1
MM1 pKa = 7.24GSRR4 pKa = 11.84YY5 pKa = 10.46AFVGAAAGARR15 pKa = 11.84QWASKK20 pKa = 10.53RR21 pKa = 11.84CVTTALKK28 pKa = 10.88YY29 pKa = 10.41ILFRR33 pKa = 11.84NNRR36 pKa = 11.84AKK38 pKa = 11.18GNDD41 pKa = 3.38CQINLTEE48 pKa = 4.61RR49 pKa = 11.84GAFVDD54 pKa = 4.09RR55 pKa = 11.84LSKK58 pKa = 10.97ADD60 pKa = 3.28ARR62 pKa = 11.84LNNTRR67 pKa = 11.84FPQLVGLTAAPLALVSSLAVSDD89 pKa = 4.77KK90 pKa = 10.95II91 pKa = 4.64
MM1 pKa = 7.24GSRR4 pKa = 11.84YY5 pKa = 10.46AFVGAAAGARR15 pKa = 11.84QWASKK20 pKa = 10.53RR21 pKa = 11.84CVTTALKK28 pKa = 10.88YY29 pKa = 10.41ILFRR33 pKa = 11.84NNRR36 pKa = 11.84AKK38 pKa = 11.18GNDD41 pKa = 3.38CQINLTEE48 pKa = 4.61RR49 pKa = 11.84GAFVDD54 pKa = 4.09RR55 pKa = 11.84LSKK58 pKa = 10.97ADD60 pKa = 3.28ARR62 pKa = 11.84LNNTRR67 pKa = 11.84FPQLVGLTAAPLALVSSLAVSDD89 pKa = 4.77KK90 pKa = 10.95II91 pKa = 4.64
Molecular weight: 9.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1497538 |
38 |
16096 |
401.5 |
44.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.817 ± 0.046 | 0.887 ± 0.028 |
6.463 ± 0.092 | 6.234 ± 0.038 |
4.464 ± 0.034 | 8.079 ± 0.065 |
1.696 ± 0.039 | 5.487 ± 0.031 |
3.917 ± 0.08 | 9.682 ± 0.09 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.994 ± 0.027 | 4.458 ± 0.041 |
4.57 ± 0.035 | 3.548 ± 0.025 |
5.527 ± 0.077 | 5.822 ± 0.032 |
6.591 ± 0.122 | 6.932 ± 0.053 |
1.202 ± 0.021 | 3.63 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
