
Aquirufa antheringensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Aquirufa
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
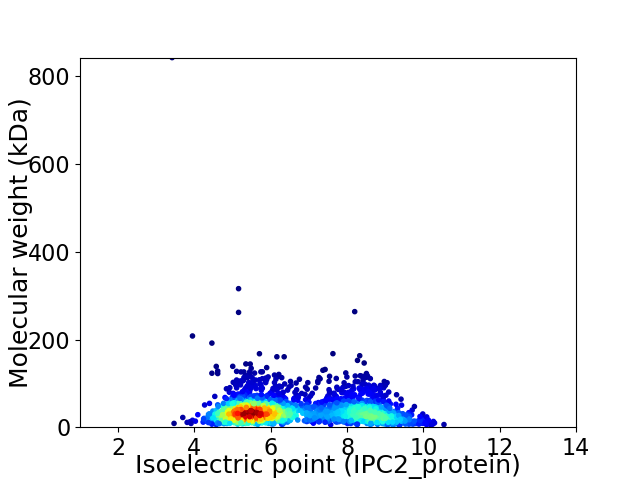
Virtual 2D-PAGE plot for 2203 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q9BHI1|A0A4Q9BHI1_9BACT LamG domain-containing protein OS=Aquirufa antheringensis OX=2516559 GN=EWU20_01575 PE=4 SV=1
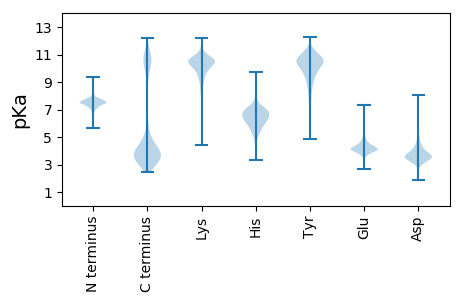
MM1 pKa = 7.85SILNNWKK8 pKa = 10.2GFLKK12 pKa = 10.53SFQKK16 pKa = 7.8TTSFLILVVSLILFSGNAHH35 pKa = 6.43AAKK38 pKa = 10.38LYY40 pKa = 11.04FNDD43 pKa = 3.38IYY45 pKa = 11.05KK46 pKa = 9.29ATGSSYY52 pKa = 10.86SIQPSSLDD60 pKa = 3.82GISFISGSGFKK71 pKa = 9.25FTSANPADD79 pKa = 3.88VSFSGNNIAGILSYY93 pKa = 10.74TNSSNQQVSIYY104 pKa = 10.87GVISRR109 pKa = 11.84QNKK112 pKa = 9.27ANGNTMAVNFMPTDD126 pKa = 3.43NTYY129 pKa = 8.2TTVTGEE135 pKa = 4.27GYY137 pKa = 10.43ILVIPSKK144 pKa = 10.65EE145 pKa = 3.75GSFQNGINVATSSDD159 pKa = 4.02PIAAVLNDD167 pKa = 3.37VLTTQNSSPIISINDD182 pKa = 3.16VTVNNTDD189 pKa = 3.0SYY191 pKa = 11.83AVFAITLSRR200 pKa = 11.84TATGITTFTPSLSPATATLNTDD222 pKa = 3.46YY223 pKa = 11.05TNSLQYY229 pKa = 11.1YY230 pKa = 9.63NGSAWVNISSTVSIAQGVTSLQIRR254 pKa = 11.84VPILNAGAVSSRR266 pKa = 11.84KK267 pKa = 9.39FNLNTGAISGSNVLNSDD284 pKa = 3.37GAYY287 pKa = 10.37GIGTILPSPSITTSGTLKK305 pKa = 10.11TFSTCSGCTVNPQAFTVSGSNLTADD330 pKa = 4.41LVVTAPTGVEE340 pKa = 3.92VSTSQNGAYY349 pKa = 9.55QSSVTLSQVSNAVTTTTVYY368 pKa = 10.55TKK370 pKa = 9.62LTNNAVSTSGGTISITSTGAVSKK393 pKa = 9.51TLTVTTNPDD402 pKa = 2.84NALNFDD408 pKa = 3.74GSSDD412 pKa = 3.96YY413 pKa = 11.63VSVPDD418 pKa = 4.19NNKK421 pKa = 10.05LDD423 pKa = 3.82FTTSFTVEE431 pKa = 3.47GWVYY435 pKa = 8.23PTTASSTQVIFGKK448 pKa = 10.59INDD451 pKa = 4.26AGSGDD456 pKa = 3.83GSDD459 pKa = 3.39AALMLRR465 pKa = 11.84LFSGGFRR472 pKa = 11.84VEE474 pKa = 4.23VGKK477 pKa = 8.71GTSPSQFFDD486 pKa = 3.53VNKK489 pKa = 9.14YY490 pKa = 7.47TLNKK494 pKa = 7.41WQHH497 pKa = 5.07IAMVYY502 pKa = 8.28NTGILKK508 pKa = 9.87FYY510 pKa = 10.53INGVEE515 pKa = 3.84EE516 pKa = 4.92GSFNTGYY523 pKa = 11.23GSTPLLNTPSSFKK536 pKa = 10.92FGVFKK541 pKa = 10.37PAFAQYY547 pKa = 10.62FIGSLDD553 pKa = 3.51NFRR556 pKa = 11.84IWDD559 pKa = 3.44IARR562 pKa = 11.84TQSEE566 pKa = 4.32ISSKK570 pKa = 10.26MNEE573 pKa = 4.15EE574 pKa = 4.74LIGTQTGLVASYY586 pKa = 9.19TFNQGIGNGNNSSITTIEE604 pKa = 4.03DD605 pKa = 3.11QTNNGLNGVLNTFSLSGSTSNFISGIIPEE634 pKa = 4.33ISGEE638 pKa = 4.21AILSKK643 pKa = 10.84GATTNYY649 pKa = 9.99TNGLTGGTWSSTDD662 pKa = 3.39TNIATVDD669 pKa = 3.49PTTGVVTGVNAGTSTITYY687 pKa = 6.91TLCDD691 pKa = 3.41KK692 pKa = 10.12TVSKK696 pKa = 10.71LVTVVIPTITTSGSLTTFTTCLGTASVAQTFTVSAQYY733 pKa = 8.95LTANLVLTAPAGYY746 pKa = 9.8EE747 pKa = 3.84LAASSGGTYY756 pKa = 10.58SSTLSIAPSSGSVSARR772 pKa = 11.84LIYY775 pKa = 10.6VRR777 pKa = 11.84LSSASIDD784 pKa = 3.88GQSGTISITSSNAVTQTITTGNASVTSSVGGSISGSASVCSGTNSTVLTLSGEE837 pKa = 4.38TGSITKK843 pKa = 8.37WQSSLTSNFTSVTDD857 pKa = 3.67IASTSASITATNLTATTYY875 pKa = 9.68YY876 pKa = 10.19RR877 pKa = 11.84AVVTLGSCTAATSSVATVTVNALPTVSVNVIPDD910 pKa = 3.41VMTSATSFTVPYY922 pKa = 10.26TNLTVGADD930 pKa = 3.76QYY932 pKa = 11.75SLTAGTTAMTNFVVVSNASVTTSPLTISIPASAQNSYY969 pKa = 11.36DD970 pKa = 3.42FNLTVKK976 pKa = 10.65NSTTGCTSAIVPFVLSVSDD995 pKa = 3.51ITPGTVSGDD1004 pKa = 3.28QSICYY1009 pKa = 9.3GSSPATITSLTDD1021 pKa = 3.29ASGGGSPTYY1030 pKa = 9.93IWQISTNGHH1039 pKa = 6.02SGTYY1043 pKa = 9.01MDD1045 pKa = 5.71IIGATSASYY1054 pKa = 10.05SHH1056 pKa = 7.13PVALTSTTHH1065 pKa = 5.74FRR1067 pKa = 11.84RR1068 pKa = 11.84KK1069 pKa = 8.57ATIGVSFAVTHH1080 pKa = 5.49PVEE1083 pKa = 4.34VLVNALPTIAVSPSSASITTGSSVSLSASGANSYY1117 pKa = 10.44SWTPSTGLSAANTAAVTATPTVTTTYY1143 pKa = 10.16TVTGTNSTTSCSSAATVTVSVNPALTAGTISADD1176 pKa = 3.25QTICVGASPATISSATAATGGTGSIAYY1203 pKa = 8.64VWSSSTNGTTYY1214 pKa = 8.26TTISGATSSTYY1225 pKa = 10.05SPGVISVNTYY1235 pKa = 7.88YY1236 pKa = 10.9KK1237 pKa = 10.44RR1238 pKa = 11.84GASTANDD1245 pKa = 3.2AVIYY1249 pKa = 7.14TSPILITVQSSVGGTIAGSAAVCAGTNSTVLTLSGYY1285 pKa = 8.16TGSVTRR1291 pKa = 11.84WQSALAADD1299 pKa = 4.68FSGIVTNINSVSTSITEE1316 pKa = 4.35TNLTATKK1323 pKa = 10.05YY1324 pKa = 8.38YY1325 pKa = 9.8RR1326 pKa = 11.84AVVRR1330 pKa = 11.84VGVCAASNSATATITVNALPVISISPSAPSITSGSSVTLTASGASTYY1377 pKa = 8.91TWTPSTGLSATNTAVVTASPTVTTTYY1403 pKa = 10.48SVSGTSSSCSSTATVTVTVTTPTPVADD1430 pKa = 3.72TDD1432 pKa = 4.01GDD1434 pKa = 4.36GVTDD1438 pKa = 3.79AQEE1441 pKa = 5.31AIDD1444 pKa = 3.96GTNPNDD1450 pKa = 3.23VCSYY1454 pKa = 10.58LVASQTLTPTSAWSTADD1471 pKa = 3.55CDD1473 pKa = 4.73GDD1475 pKa = 3.72GTPNVTDD1482 pKa = 4.08SDD1484 pKa = 4.39PNDD1487 pKa = 3.94PCVHH1491 pKa = 6.84LAGATPVTTNAIWRR1505 pKa = 11.84AADD1508 pKa = 3.56CDD1510 pKa = 4.09GDD1512 pKa = 3.99GVTNGKK1518 pKa = 8.37EE1519 pKa = 4.28TDD1521 pKa = 4.37DD1522 pKa = 3.74GTDD1525 pKa = 3.47PNEE1528 pKa = 4.16GCSYY1532 pKa = 10.57VVASRR1537 pKa = 11.84TLTPSTAWGTLDD1549 pKa = 4.78CDD1551 pKa = 4.51ADD1553 pKa = 4.12GNSNATDD1560 pKa = 4.07PNIDD1564 pKa = 3.92APVASNDD1571 pKa = 3.58LVNLPSSGALTVNILTNDD1589 pKa = 3.56DD1590 pKa = 4.37FLPGANTEE1598 pKa = 4.24ISRR1601 pKa = 11.84QLPPNDD1607 pKa = 3.32GTAKK1611 pKa = 9.56GTVTFNPLTGEE1622 pKa = 4.09MTYY1625 pKa = 10.87KK1626 pKa = 10.54RR1627 pKa = 11.84APFEE1631 pKa = 4.28TGLVTMGYY1639 pKa = 10.15KK1640 pKa = 8.68VTNKK1644 pKa = 9.91AVSPPVSAFAFVTILACDD1662 pKa = 3.8LQDD1665 pKa = 4.76PLSDD1669 pKa = 3.87CDD1671 pKa = 5.71GDD1673 pKa = 4.22GEE1675 pKa = 5.03PNGTDD1680 pKa = 3.82LAPTDD1685 pKa = 3.62PCVYY1689 pKa = 10.28EE1690 pKa = 3.95PTRR1693 pKa = 11.84QVKK1696 pKa = 10.69ANVSDD1701 pKa = 3.39DD1702 pKa = 3.62WKK1704 pKa = 11.46ALDD1707 pKa = 4.1CDD1709 pKa = 4.04GDD1711 pKa = 4.12GVLNGTEE1718 pKa = 4.17LADD1721 pKa = 3.7GTSPSDD1727 pKa = 3.18ACSFKK1732 pKa = 10.92AGSITLSPSAAWLLLDD1748 pKa = 4.77CDD1750 pKa = 4.32ADD1752 pKa = 3.81GVTNQVEE1759 pKa = 4.66GTQDD1763 pKa = 3.47LDD1765 pKa = 3.61GDD1767 pKa = 4.95GIPNCLDD1774 pKa = 3.25TDD1776 pKa = 3.81SDD1778 pKa = 4.18GDD1780 pKa = 4.34SISDD1784 pKa = 3.54KK1785 pKa = 11.3LEE1787 pKa = 4.4TIVDD1791 pKa = 3.59TDD1793 pKa = 3.71RR1794 pKa = 11.84DD1795 pKa = 4.08GKK1797 pKa = 9.95PDD1799 pKa = 3.9YY1800 pKa = 11.22LDD1802 pKa = 5.15LDD1804 pKa = 4.0SDD1806 pKa = 3.94NDD1808 pKa = 4.66GILDD1812 pKa = 3.67SVEE1815 pKa = 4.5NKK1817 pKa = 9.64VCTGTGILCDD1827 pKa = 3.72TDD1829 pKa = 5.23ADD1831 pKa = 3.95GTPNFRR1837 pKa = 11.84DD1838 pKa = 3.58LDD1840 pKa = 3.89SDD1842 pKa = 3.74ADD1844 pKa = 3.83QIKK1847 pKa = 10.69DD1848 pKa = 3.59VIEE1851 pKa = 4.52ASGSDD1856 pKa = 3.71YY1857 pKa = 11.25NQDD1860 pKa = 3.45GIADD1864 pKa = 3.84GNIDD1868 pKa = 3.66EE1869 pKa = 5.1KK1870 pKa = 11.34GIPSSANKK1878 pKa = 10.33GLTPPDD1884 pKa = 3.37TDD1886 pKa = 4.19RR1887 pKa = 11.84DD1888 pKa = 4.1GKK1890 pKa = 10.83LDD1892 pKa = 4.19PYY1894 pKa = 11.3DD1895 pKa = 3.78VDD1897 pKa = 4.07SDD1899 pKa = 3.99GDD1901 pKa = 4.18GISDD1905 pKa = 3.84FDD1907 pKa = 3.7EE1908 pKa = 5.17EE1909 pKa = 4.61YY1910 pKa = 11.54NEE1912 pKa = 4.12VADD1915 pKa = 6.35FADD1918 pKa = 4.11CDD1920 pKa = 3.56KK1921 pKa = 11.6DD1922 pKa = 4.58GIVNRR1927 pKa = 11.84LDD1929 pKa = 3.74PDD1931 pKa = 3.33EE1932 pKa = 5.71CEE1934 pKa = 3.76IFAPQGISPNNDD1946 pKa = 3.12DD1947 pKa = 4.18KK1948 pKa = 11.8NDD1950 pKa = 3.69RR1951 pKa = 11.84LVFKK1955 pKa = 10.8GLVFRR1960 pKa = 11.84KK1961 pKa = 9.86LPNHH1965 pKa = 6.33LSVYY1969 pKa = 9.67NRR1971 pKa = 11.84WGTLVFEE1978 pKa = 4.79MDD1980 pKa = 4.95DD1981 pKa = 4.2YY1982 pKa = 12.03DD1983 pKa = 4.26NSWSGTILPDD1993 pKa = 3.05GTYY1996 pKa = 10.25FYY1998 pKa = 11.7VLDD2001 pKa = 4.46FYY2003 pKa = 11.5GRR2005 pKa = 11.84KK2006 pKa = 7.8PTISNYY2012 pKa = 10.11LAIDD2016 pKa = 3.57RR2017 pKa = 11.84TIKK2020 pKa = 10.68
MM1 pKa = 7.85SILNNWKK8 pKa = 10.2GFLKK12 pKa = 10.53SFQKK16 pKa = 7.8TTSFLILVVSLILFSGNAHH35 pKa = 6.43AAKK38 pKa = 10.38LYY40 pKa = 11.04FNDD43 pKa = 3.38IYY45 pKa = 11.05KK46 pKa = 9.29ATGSSYY52 pKa = 10.86SIQPSSLDD60 pKa = 3.82GISFISGSGFKK71 pKa = 9.25FTSANPADD79 pKa = 3.88VSFSGNNIAGILSYY93 pKa = 10.74TNSSNQQVSIYY104 pKa = 10.87GVISRR109 pKa = 11.84QNKK112 pKa = 9.27ANGNTMAVNFMPTDD126 pKa = 3.43NTYY129 pKa = 8.2TTVTGEE135 pKa = 4.27GYY137 pKa = 10.43ILVIPSKK144 pKa = 10.65EE145 pKa = 3.75GSFQNGINVATSSDD159 pKa = 4.02PIAAVLNDD167 pKa = 3.37VLTTQNSSPIISINDD182 pKa = 3.16VTVNNTDD189 pKa = 3.0SYY191 pKa = 11.83AVFAITLSRR200 pKa = 11.84TATGITTFTPSLSPATATLNTDD222 pKa = 3.46YY223 pKa = 11.05TNSLQYY229 pKa = 11.1YY230 pKa = 9.63NGSAWVNISSTVSIAQGVTSLQIRR254 pKa = 11.84VPILNAGAVSSRR266 pKa = 11.84KK267 pKa = 9.39FNLNTGAISGSNVLNSDD284 pKa = 3.37GAYY287 pKa = 10.37GIGTILPSPSITTSGTLKK305 pKa = 10.11TFSTCSGCTVNPQAFTVSGSNLTADD330 pKa = 4.41LVVTAPTGVEE340 pKa = 3.92VSTSQNGAYY349 pKa = 9.55QSSVTLSQVSNAVTTTTVYY368 pKa = 10.55TKK370 pKa = 9.62LTNNAVSTSGGTISITSTGAVSKK393 pKa = 9.51TLTVTTNPDD402 pKa = 2.84NALNFDD408 pKa = 3.74GSSDD412 pKa = 3.96YY413 pKa = 11.63VSVPDD418 pKa = 4.19NNKK421 pKa = 10.05LDD423 pKa = 3.82FTTSFTVEE431 pKa = 3.47GWVYY435 pKa = 8.23PTTASSTQVIFGKK448 pKa = 10.59INDD451 pKa = 4.26AGSGDD456 pKa = 3.83GSDD459 pKa = 3.39AALMLRR465 pKa = 11.84LFSGGFRR472 pKa = 11.84VEE474 pKa = 4.23VGKK477 pKa = 8.71GTSPSQFFDD486 pKa = 3.53VNKK489 pKa = 9.14YY490 pKa = 7.47TLNKK494 pKa = 7.41WQHH497 pKa = 5.07IAMVYY502 pKa = 8.28NTGILKK508 pKa = 9.87FYY510 pKa = 10.53INGVEE515 pKa = 3.84EE516 pKa = 4.92GSFNTGYY523 pKa = 11.23GSTPLLNTPSSFKK536 pKa = 10.92FGVFKK541 pKa = 10.37PAFAQYY547 pKa = 10.62FIGSLDD553 pKa = 3.51NFRR556 pKa = 11.84IWDD559 pKa = 3.44IARR562 pKa = 11.84TQSEE566 pKa = 4.32ISSKK570 pKa = 10.26MNEE573 pKa = 4.15EE574 pKa = 4.74LIGTQTGLVASYY586 pKa = 9.19TFNQGIGNGNNSSITTIEE604 pKa = 4.03DD605 pKa = 3.11QTNNGLNGVLNTFSLSGSTSNFISGIIPEE634 pKa = 4.33ISGEE638 pKa = 4.21AILSKK643 pKa = 10.84GATTNYY649 pKa = 9.99TNGLTGGTWSSTDD662 pKa = 3.39TNIATVDD669 pKa = 3.49PTTGVVTGVNAGTSTITYY687 pKa = 6.91TLCDD691 pKa = 3.41KK692 pKa = 10.12TVSKK696 pKa = 10.71LVTVVIPTITTSGSLTTFTTCLGTASVAQTFTVSAQYY733 pKa = 8.95LTANLVLTAPAGYY746 pKa = 9.8EE747 pKa = 3.84LAASSGGTYY756 pKa = 10.58SSTLSIAPSSGSVSARR772 pKa = 11.84LIYY775 pKa = 10.6VRR777 pKa = 11.84LSSASIDD784 pKa = 3.88GQSGTISITSSNAVTQTITTGNASVTSSVGGSISGSASVCSGTNSTVLTLSGEE837 pKa = 4.38TGSITKK843 pKa = 8.37WQSSLTSNFTSVTDD857 pKa = 3.67IASTSASITATNLTATTYY875 pKa = 9.68YY876 pKa = 10.19RR877 pKa = 11.84AVVTLGSCTAATSSVATVTVNALPTVSVNVIPDD910 pKa = 3.41VMTSATSFTVPYY922 pKa = 10.26TNLTVGADD930 pKa = 3.76QYY932 pKa = 11.75SLTAGTTAMTNFVVVSNASVTTSPLTISIPASAQNSYY969 pKa = 11.36DD970 pKa = 3.42FNLTVKK976 pKa = 10.65NSTTGCTSAIVPFVLSVSDD995 pKa = 3.51ITPGTVSGDD1004 pKa = 3.28QSICYY1009 pKa = 9.3GSSPATITSLTDD1021 pKa = 3.29ASGGGSPTYY1030 pKa = 9.93IWQISTNGHH1039 pKa = 6.02SGTYY1043 pKa = 9.01MDD1045 pKa = 5.71IIGATSASYY1054 pKa = 10.05SHH1056 pKa = 7.13PVALTSTTHH1065 pKa = 5.74FRR1067 pKa = 11.84RR1068 pKa = 11.84KK1069 pKa = 8.57ATIGVSFAVTHH1080 pKa = 5.49PVEE1083 pKa = 4.34VLVNALPTIAVSPSSASITTGSSVSLSASGANSYY1117 pKa = 10.44SWTPSTGLSAANTAAVTATPTVTTTYY1143 pKa = 10.16TVTGTNSTTSCSSAATVTVSVNPALTAGTISADD1176 pKa = 3.25QTICVGASPATISSATAATGGTGSIAYY1203 pKa = 8.64VWSSSTNGTTYY1214 pKa = 8.26TTISGATSSTYY1225 pKa = 10.05SPGVISVNTYY1235 pKa = 7.88YY1236 pKa = 10.9KK1237 pKa = 10.44RR1238 pKa = 11.84GASTANDD1245 pKa = 3.2AVIYY1249 pKa = 7.14TSPILITVQSSVGGTIAGSAAVCAGTNSTVLTLSGYY1285 pKa = 8.16TGSVTRR1291 pKa = 11.84WQSALAADD1299 pKa = 4.68FSGIVTNINSVSTSITEE1316 pKa = 4.35TNLTATKK1323 pKa = 10.05YY1324 pKa = 8.38YY1325 pKa = 9.8RR1326 pKa = 11.84AVVRR1330 pKa = 11.84VGVCAASNSATATITVNALPVISISPSAPSITSGSSVTLTASGASTYY1377 pKa = 8.91TWTPSTGLSATNTAVVTASPTVTTTYY1403 pKa = 10.48SVSGTSSSCSSTATVTVTVTTPTPVADD1430 pKa = 3.72TDD1432 pKa = 4.01GDD1434 pKa = 4.36GVTDD1438 pKa = 3.79AQEE1441 pKa = 5.31AIDD1444 pKa = 3.96GTNPNDD1450 pKa = 3.23VCSYY1454 pKa = 10.58LVASQTLTPTSAWSTADD1471 pKa = 3.55CDD1473 pKa = 4.73GDD1475 pKa = 3.72GTPNVTDD1482 pKa = 4.08SDD1484 pKa = 4.39PNDD1487 pKa = 3.94PCVHH1491 pKa = 6.84LAGATPVTTNAIWRR1505 pKa = 11.84AADD1508 pKa = 3.56CDD1510 pKa = 4.09GDD1512 pKa = 3.99GVTNGKK1518 pKa = 8.37EE1519 pKa = 4.28TDD1521 pKa = 4.37DD1522 pKa = 3.74GTDD1525 pKa = 3.47PNEE1528 pKa = 4.16GCSYY1532 pKa = 10.57VVASRR1537 pKa = 11.84TLTPSTAWGTLDD1549 pKa = 4.78CDD1551 pKa = 4.51ADD1553 pKa = 4.12GNSNATDD1560 pKa = 4.07PNIDD1564 pKa = 3.92APVASNDD1571 pKa = 3.58LVNLPSSGALTVNILTNDD1589 pKa = 3.56DD1590 pKa = 4.37FLPGANTEE1598 pKa = 4.24ISRR1601 pKa = 11.84QLPPNDD1607 pKa = 3.32GTAKK1611 pKa = 9.56GTVTFNPLTGEE1622 pKa = 4.09MTYY1625 pKa = 10.87KK1626 pKa = 10.54RR1627 pKa = 11.84APFEE1631 pKa = 4.28TGLVTMGYY1639 pKa = 10.15KK1640 pKa = 8.68VTNKK1644 pKa = 9.91AVSPPVSAFAFVTILACDD1662 pKa = 3.8LQDD1665 pKa = 4.76PLSDD1669 pKa = 3.87CDD1671 pKa = 5.71GDD1673 pKa = 4.22GEE1675 pKa = 5.03PNGTDD1680 pKa = 3.82LAPTDD1685 pKa = 3.62PCVYY1689 pKa = 10.28EE1690 pKa = 3.95PTRR1693 pKa = 11.84QVKK1696 pKa = 10.69ANVSDD1701 pKa = 3.39DD1702 pKa = 3.62WKK1704 pKa = 11.46ALDD1707 pKa = 4.1CDD1709 pKa = 4.04GDD1711 pKa = 4.12GVLNGTEE1718 pKa = 4.17LADD1721 pKa = 3.7GTSPSDD1727 pKa = 3.18ACSFKK1732 pKa = 10.92AGSITLSPSAAWLLLDD1748 pKa = 4.77CDD1750 pKa = 4.32ADD1752 pKa = 3.81GVTNQVEE1759 pKa = 4.66GTQDD1763 pKa = 3.47LDD1765 pKa = 3.61GDD1767 pKa = 4.95GIPNCLDD1774 pKa = 3.25TDD1776 pKa = 3.81SDD1778 pKa = 4.18GDD1780 pKa = 4.34SISDD1784 pKa = 3.54KK1785 pKa = 11.3LEE1787 pKa = 4.4TIVDD1791 pKa = 3.59TDD1793 pKa = 3.71RR1794 pKa = 11.84DD1795 pKa = 4.08GKK1797 pKa = 9.95PDD1799 pKa = 3.9YY1800 pKa = 11.22LDD1802 pKa = 5.15LDD1804 pKa = 4.0SDD1806 pKa = 3.94NDD1808 pKa = 4.66GILDD1812 pKa = 3.67SVEE1815 pKa = 4.5NKK1817 pKa = 9.64VCTGTGILCDD1827 pKa = 3.72TDD1829 pKa = 5.23ADD1831 pKa = 3.95GTPNFRR1837 pKa = 11.84DD1838 pKa = 3.58LDD1840 pKa = 3.89SDD1842 pKa = 3.74ADD1844 pKa = 3.83QIKK1847 pKa = 10.69DD1848 pKa = 3.59VIEE1851 pKa = 4.52ASGSDD1856 pKa = 3.71YY1857 pKa = 11.25NQDD1860 pKa = 3.45GIADD1864 pKa = 3.84GNIDD1868 pKa = 3.66EE1869 pKa = 5.1KK1870 pKa = 11.34GIPSSANKK1878 pKa = 10.33GLTPPDD1884 pKa = 3.37TDD1886 pKa = 4.19RR1887 pKa = 11.84DD1888 pKa = 4.1GKK1890 pKa = 10.83LDD1892 pKa = 4.19PYY1894 pKa = 11.3DD1895 pKa = 3.78VDD1897 pKa = 4.07SDD1899 pKa = 3.99GDD1901 pKa = 4.18GISDD1905 pKa = 3.84FDD1907 pKa = 3.7EE1908 pKa = 5.17EE1909 pKa = 4.61YY1910 pKa = 11.54NEE1912 pKa = 4.12VADD1915 pKa = 6.35FADD1918 pKa = 4.11CDD1920 pKa = 3.56KK1921 pKa = 11.6DD1922 pKa = 4.58GIVNRR1927 pKa = 11.84LDD1929 pKa = 3.74PDD1931 pKa = 3.33EE1932 pKa = 5.71CEE1934 pKa = 3.76IFAPQGISPNNDD1946 pKa = 3.12DD1947 pKa = 4.18KK1948 pKa = 11.8NDD1950 pKa = 3.69RR1951 pKa = 11.84LVFKK1955 pKa = 10.8GLVFRR1960 pKa = 11.84KK1961 pKa = 9.86LPNHH1965 pKa = 6.33LSVYY1969 pKa = 9.67NRR1971 pKa = 11.84WGTLVFEE1978 pKa = 4.79MDD1980 pKa = 4.95DD1981 pKa = 4.2YY1982 pKa = 12.03DD1983 pKa = 4.26NSWSGTILPDD1993 pKa = 3.05GTYY1996 pKa = 10.25FYY1998 pKa = 11.7VLDD2001 pKa = 4.46FYY2003 pKa = 11.5GRR2005 pKa = 11.84KK2006 pKa = 7.8PTISNYY2012 pKa = 10.11LAIDD2016 pKa = 3.57RR2017 pKa = 11.84TIKK2020 pKa = 10.68
Molecular weight: 208.04 kDa
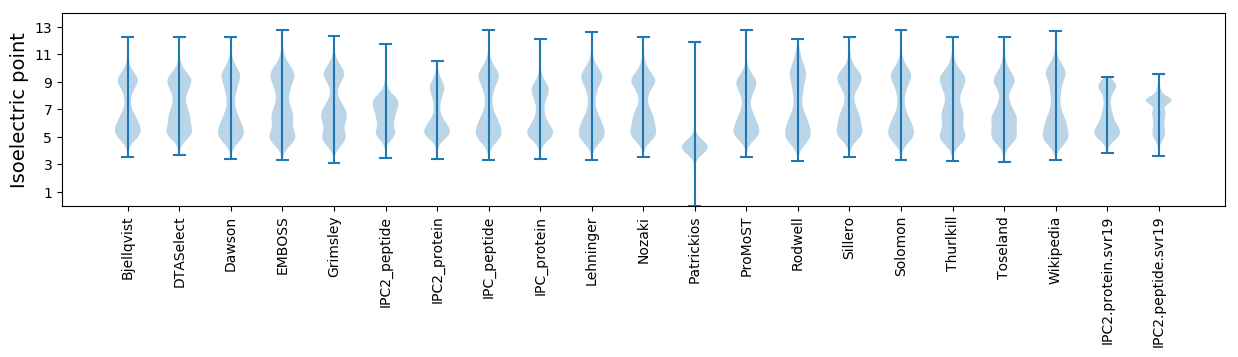
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q9BA72|A0A4Q9BA72_9BACT DNA alkylation repair protein OS=Aquirufa antheringensis OX=2516559 GN=EWU20_09965 PE=4 SV=1
MM1 pKa = 7.41NPTNAKK7 pKa = 9.66NRR9 pKa = 11.84LFLAVGLVGTLCLLSACWSARR30 pKa = 11.84CPYY33 pKa = 7.46QTCRR37 pKa = 11.84VKK39 pKa = 10.52VEE41 pKa = 3.89HH42 pKa = 6.23RR43 pKa = 11.84HH44 pKa = 4.23GVGYY48 pKa = 9.45FRR50 pKa = 11.84PNVAFSWMWTPRR62 pKa = 11.84YY63 pKa = 9.74KK64 pKa = 10.47HH65 pKa = 6.04IRR67 pKa = 11.84ADD69 pKa = 3.53QSKK72 pKa = 10.28KK73 pKa = 10.36KK74 pKa = 10.15PRR76 pKa = 3.57
MM1 pKa = 7.41NPTNAKK7 pKa = 9.66NRR9 pKa = 11.84LFLAVGLVGTLCLLSACWSARR30 pKa = 11.84CPYY33 pKa = 7.46QTCRR37 pKa = 11.84VKK39 pKa = 10.52VEE41 pKa = 3.89HH42 pKa = 6.23RR43 pKa = 11.84HH44 pKa = 4.23GVGYY48 pKa = 9.45FRR50 pKa = 11.84PNVAFSWMWTPRR62 pKa = 11.84YY63 pKa = 9.74KK64 pKa = 10.47HH65 pKa = 6.04IRR67 pKa = 11.84ADD69 pKa = 3.53QSKK72 pKa = 10.28KK73 pKa = 10.36KK74 pKa = 10.15PRR76 pKa = 3.57
Molecular weight: 8.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
789050 |
31 |
8145 |
358.2 |
39.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.125 ± 0.054 | 0.774 ± 0.015 |
5.091 ± 0.066 | 6.033 ± 0.066 |
5.067 ± 0.044 | 7.247 ± 0.056 |
1.873 ± 0.026 | 6.949 ± 0.043 |
6.705 ± 0.066 | 9.691 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.368 ± 0.031 | 4.754 ± 0.054 |
4.017 ± 0.03 | 3.757 ± 0.036 |
3.806 ± 0.04 | 6.68 ± 0.056 |
5.634 ± 0.102 | 6.505 ± 0.036 |
1.209 ± 0.02 | 3.715 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
