
Sanxia tombus-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.77
Get precalculated fractions of proteins
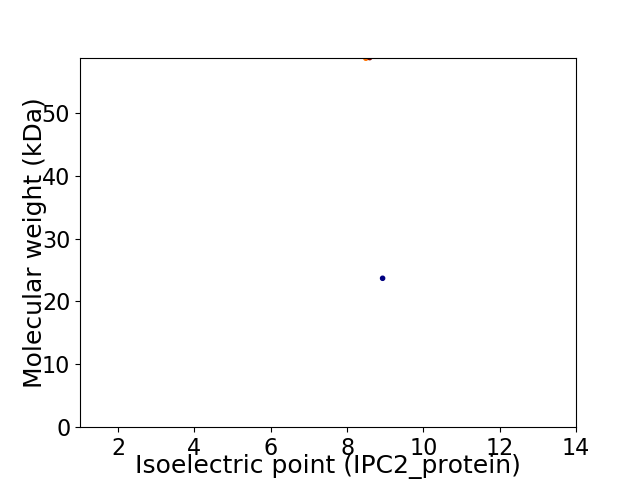
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGL5|A0A1L3KGL5_9VIRU RNA-directed RNA polymerase OS=Sanxia tombus-like virus 3 OX=1923387 PE=4 SV=1
MM1 pKa = 7.62LEE3 pKa = 4.32GVDD6 pKa = 4.08TVPGVYY12 pKa = 9.95PGISPTKK19 pKa = 10.54LPGIPKK25 pKa = 8.83NRR27 pKa = 11.84KK28 pKa = 7.58YY29 pKa = 10.66LTMGQYY35 pKa = 9.58ICDD38 pKa = 3.82YY39 pKa = 10.53EE40 pKa = 4.53VRR42 pKa = 11.84THH44 pKa = 6.77NNSLNNLIRR53 pKa = 11.84GVGEE57 pKa = 3.72RR58 pKa = 11.84VLYY61 pKa = 9.56TDD63 pKa = 5.01KK64 pKa = 11.52NCTPCIKK71 pKa = 9.75PARR74 pKa = 11.84GGIFRR79 pKa = 11.84DD80 pKa = 3.13RR81 pKa = 11.84CGVYY85 pKa = 9.37MRR87 pKa = 11.84TIAQRR92 pKa = 11.84LGHH95 pKa = 5.87QSPVSRR101 pKa = 11.84QQFVEE106 pKa = 5.32FYY108 pKa = 9.45SGRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84AMYY116 pKa = 9.97QKK118 pKa = 10.85AVDD121 pKa = 3.95GLALKK126 pKa = 9.58PIRR129 pKa = 11.84VLDD132 pKa = 3.63SHH134 pKa = 7.15LSTFIKK140 pKa = 10.48AEE142 pKa = 3.91KK143 pKa = 10.42LIFTLKK149 pKa = 9.97PDD151 pKa = 3.98PAPRR155 pKa = 11.84VIQPRR160 pKa = 11.84RR161 pKa = 11.84PVYY164 pKa = 9.43NVEE167 pKa = 3.61VGKK170 pKa = 9.32YY171 pKa = 9.34LRR173 pKa = 11.84PLEE176 pKa = 3.99SRR178 pKa = 11.84IYY180 pKa = 10.95DD181 pKa = 3.87EE182 pKa = 4.53IDD184 pKa = 3.25NIFRR188 pKa = 11.84SPTIMSKK195 pKa = 10.65HH196 pKa = 5.75NSVKK200 pKa = 9.33QAEE203 pKa = 4.59IIVSKK208 pKa = 9.28WQQFQQPACVGLDD221 pKa = 3.07ASRR224 pKa = 11.84FDD226 pKa = 3.47QHH228 pKa = 8.35VSEE231 pKa = 4.21QALKK235 pKa = 10.69FEE237 pKa = 4.63HH238 pKa = 6.58EE239 pKa = 4.77LYY241 pKa = 11.14NSVFKK246 pKa = 10.72CKK248 pKa = 10.1RR249 pKa = 11.84LAKK252 pKa = 10.47LLTWQINNVGHH263 pKa = 7.03ARR265 pKa = 11.84AADD268 pKa = 3.55GKK270 pKa = 9.2FTYY273 pKa = 8.67RR274 pKa = 11.84TKK276 pKa = 10.61GSRR279 pKa = 11.84MSGDD283 pKa = 3.39MNTSLGNKK291 pKa = 9.44FLMCLMCKK299 pKa = 10.19AYY301 pKa = 9.81IDD303 pKa = 3.68TKK305 pKa = 10.42RR306 pKa = 11.84IKK308 pKa = 10.09IDD310 pKa = 3.58FVNNGDD316 pKa = 3.75DD317 pKa = 3.54CLMFLEE323 pKa = 5.14RR324 pKa = 11.84KK325 pKa = 9.4HH326 pKa = 6.81LSQLHH331 pKa = 5.54NLQSYY336 pKa = 8.84FADD339 pKa = 4.06FGFKK343 pKa = 9.37MVTEE347 pKa = 4.34APVFEE352 pKa = 4.89LEE354 pKa = 5.04HH355 pKa = 8.77IEE357 pKa = 4.31FCQSRR362 pKa = 11.84PLKK365 pKa = 11.0CNGIWRR371 pKa = 11.84MTRR374 pKa = 11.84NIRR377 pKa = 11.84TCLARR382 pKa = 11.84DD383 pKa = 3.46VTAVNFGHH391 pKa = 6.53DD392 pKa = 3.36VCMYY396 pKa = 10.09RR397 pKa = 11.84AWLHH401 pKa = 7.06DD402 pKa = 3.33IANCGLAFAGDD413 pKa = 4.01CPVYY417 pKa = 10.58GSFYY421 pKa = 11.11RR422 pKa = 11.84MLLRR426 pKa = 11.84FGVEE430 pKa = 3.91GNYY433 pKa = 9.86HH434 pKa = 6.7DD435 pKa = 5.15RR436 pKa = 11.84HH437 pKa = 5.85DD438 pKa = 4.36AYY440 pKa = 11.22FNVYY444 pKa = 7.97RR445 pKa = 11.84TMSRR449 pKa = 11.84GIDD452 pKa = 3.48VEE454 pKa = 4.48TNQPDD459 pKa = 3.48DD460 pKa = 3.64YY461 pKa = 11.43GRR463 pKa = 11.84YY464 pKa = 8.84SFWMQTGINPDD475 pKa = 3.12AQFEE479 pKa = 4.42LEE481 pKa = 4.21QYY483 pKa = 10.55FDD485 pKa = 3.54NAVWGGDD492 pKa = 2.76KK493 pKa = 10.69RR494 pKa = 11.84QFITNLSRR502 pKa = 11.84LITHH506 pKa = 7.11GSS508 pKa = 3.16
MM1 pKa = 7.62LEE3 pKa = 4.32GVDD6 pKa = 4.08TVPGVYY12 pKa = 9.95PGISPTKK19 pKa = 10.54LPGIPKK25 pKa = 8.83NRR27 pKa = 11.84KK28 pKa = 7.58YY29 pKa = 10.66LTMGQYY35 pKa = 9.58ICDD38 pKa = 3.82YY39 pKa = 10.53EE40 pKa = 4.53VRR42 pKa = 11.84THH44 pKa = 6.77NNSLNNLIRR53 pKa = 11.84GVGEE57 pKa = 3.72RR58 pKa = 11.84VLYY61 pKa = 9.56TDD63 pKa = 5.01KK64 pKa = 11.52NCTPCIKK71 pKa = 9.75PARR74 pKa = 11.84GGIFRR79 pKa = 11.84DD80 pKa = 3.13RR81 pKa = 11.84CGVYY85 pKa = 9.37MRR87 pKa = 11.84TIAQRR92 pKa = 11.84LGHH95 pKa = 5.87QSPVSRR101 pKa = 11.84QQFVEE106 pKa = 5.32FYY108 pKa = 9.45SGRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84AMYY116 pKa = 9.97QKK118 pKa = 10.85AVDD121 pKa = 3.95GLALKK126 pKa = 9.58PIRR129 pKa = 11.84VLDD132 pKa = 3.63SHH134 pKa = 7.15LSTFIKK140 pKa = 10.48AEE142 pKa = 3.91KK143 pKa = 10.42LIFTLKK149 pKa = 9.97PDD151 pKa = 3.98PAPRR155 pKa = 11.84VIQPRR160 pKa = 11.84RR161 pKa = 11.84PVYY164 pKa = 9.43NVEE167 pKa = 3.61VGKK170 pKa = 9.32YY171 pKa = 9.34LRR173 pKa = 11.84PLEE176 pKa = 3.99SRR178 pKa = 11.84IYY180 pKa = 10.95DD181 pKa = 3.87EE182 pKa = 4.53IDD184 pKa = 3.25NIFRR188 pKa = 11.84SPTIMSKK195 pKa = 10.65HH196 pKa = 5.75NSVKK200 pKa = 9.33QAEE203 pKa = 4.59IIVSKK208 pKa = 9.28WQQFQQPACVGLDD221 pKa = 3.07ASRR224 pKa = 11.84FDD226 pKa = 3.47QHH228 pKa = 8.35VSEE231 pKa = 4.21QALKK235 pKa = 10.69FEE237 pKa = 4.63HH238 pKa = 6.58EE239 pKa = 4.77LYY241 pKa = 11.14NSVFKK246 pKa = 10.72CKK248 pKa = 10.1RR249 pKa = 11.84LAKK252 pKa = 10.47LLTWQINNVGHH263 pKa = 7.03ARR265 pKa = 11.84AADD268 pKa = 3.55GKK270 pKa = 9.2FTYY273 pKa = 8.67RR274 pKa = 11.84TKK276 pKa = 10.61GSRR279 pKa = 11.84MSGDD283 pKa = 3.39MNTSLGNKK291 pKa = 9.44FLMCLMCKK299 pKa = 10.19AYY301 pKa = 9.81IDD303 pKa = 3.68TKK305 pKa = 10.42RR306 pKa = 11.84IKK308 pKa = 10.09IDD310 pKa = 3.58FVNNGDD316 pKa = 3.75DD317 pKa = 3.54CLMFLEE323 pKa = 5.14RR324 pKa = 11.84KK325 pKa = 9.4HH326 pKa = 6.81LSQLHH331 pKa = 5.54NLQSYY336 pKa = 8.84FADD339 pKa = 4.06FGFKK343 pKa = 9.37MVTEE347 pKa = 4.34APVFEE352 pKa = 4.89LEE354 pKa = 5.04HH355 pKa = 8.77IEE357 pKa = 4.31FCQSRR362 pKa = 11.84PLKK365 pKa = 11.0CNGIWRR371 pKa = 11.84MTRR374 pKa = 11.84NIRR377 pKa = 11.84TCLARR382 pKa = 11.84DD383 pKa = 3.46VTAVNFGHH391 pKa = 6.53DD392 pKa = 3.36VCMYY396 pKa = 10.09RR397 pKa = 11.84AWLHH401 pKa = 7.06DD402 pKa = 3.33IANCGLAFAGDD413 pKa = 4.01CPVYY417 pKa = 10.58GSFYY421 pKa = 11.11RR422 pKa = 11.84MLLRR426 pKa = 11.84FGVEE430 pKa = 3.91GNYY433 pKa = 9.86HH434 pKa = 6.7DD435 pKa = 5.15RR436 pKa = 11.84HH437 pKa = 5.85DD438 pKa = 4.36AYY440 pKa = 11.22FNVYY444 pKa = 7.97RR445 pKa = 11.84TMSRR449 pKa = 11.84GIDD452 pKa = 3.48VEE454 pKa = 4.48TNQPDD459 pKa = 3.48DD460 pKa = 3.64YY461 pKa = 11.43GRR463 pKa = 11.84YY464 pKa = 8.84SFWMQTGINPDD475 pKa = 3.12AQFEE479 pKa = 4.42LEE481 pKa = 4.21QYY483 pKa = 10.55FDD485 pKa = 3.54NAVWGGDD492 pKa = 2.76KK493 pKa = 10.69RR494 pKa = 11.84QFITNLSRR502 pKa = 11.84LITHH506 pKa = 7.11GSS508 pKa = 3.16
Molecular weight: 58.67 kDa
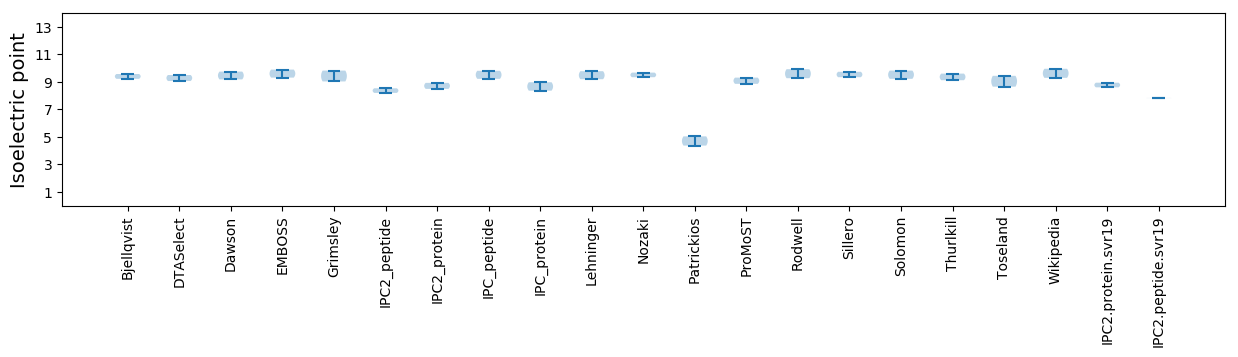
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGL5|A0A1L3KGL5_9VIRU RNA-directed RNA polymerase OS=Sanxia tombus-like virus 3 OX=1923387 PE=4 SV=1
MM1 pKa = 7.46APKK4 pKa = 10.15GKK6 pKa = 9.96KK7 pKa = 9.37VSNTVRR13 pKa = 11.84NAVDD17 pKa = 3.04SRR19 pKa = 11.84SRR21 pKa = 11.84GGIRR25 pKa = 11.84PRR27 pKa = 11.84IRR29 pKa = 11.84GTLGNNMVLRR39 pKa = 11.84GNEE42 pKa = 3.74MLEE45 pKa = 4.02NLKK48 pKa = 10.52VPATQTDD55 pKa = 3.26HH56 pKa = 6.86SLGYY60 pKa = 9.47PLIAGVTVARR70 pKa = 11.84DD71 pKa = 3.54SVIFAGVARR80 pKa = 11.84CFQQYY85 pKa = 10.51KK86 pKa = 9.19YY87 pKa = 11.2LPGTALHH94 pKa = 5.79YY95 pKa = 10.4QPSVGLNTSGTIYY108 pKa = 10.68AAWFEE113 pKa = 4.17NPEE116 pKa = 4.27MVSDD120 pKa = 3.83WDD122 pKa = 3.39ILGTVARR129 pKa = 11.84IARR132 pKa = 11.84IKK134 pKa = 10.86ANANVKK140 pKa = 9.73CFPVWQEE147 pKa = 3.1FRR149 pKa = 11.84IPISGTPRR157 pKa = 11.84QKK159 pKa = 10.13MFSTDD164 pKa = 2.14ISTNYY169 pKa = 10.1SVADD173 pKa = 3.64LTRR176 pKa = 11.84TCQGLFAVAIEE187 pKa = 4.24GLTANATAAQTIGRR201 pKa = 11.84CVLEE205 pKa = 4.12TVLQLEE211 pKa = 4.69GLSNPTSS218 pKa = 3.33
MM1 pKa = 7.46APKK4 pKa = 10.15GKK6 pKa = 9.96KK7 pKa = 9.37VSNTVRR13 pKa = 11.84NAVDD17 pKa = 3.04SRR19 pKa = 11.84SRR21 pKa = 11.84GGIRR25 pKa = 11.84PRR27 pKa = 11.84IRR29 pKa = 11.84GTLGNNMVLRR39 pKa = 11.84GNEE42 pKa = 3.74MLEE45 pKa = 4.02NLKK48 pKa = 10.52VPATQTDD55 pKa = 3.26HH56 pKa = 6.86SLGYY60 pKa = 9.47PLIAGVTVARR70 pKa = 11.84DD71 pKa = 3.54SVIFAGVARR80 pKa = 11.84CFQQYY85 pKa = 10.51KK86 pKa = 9.19YY87 pKa = 11.2LPGTALHH94 pKa = 5.79YY95 pKa = 10.4QPSVGLNTSGTIYY108 pKa = 10.68AAWFEE113 pKa = 4.17NPEE116 pKa = 4.27MVSDD120 pKa = 3.83WDD122 pKa = 3.39ILGTVARR129 pKa = 11.84IARR132 pKa = 11.84IKK134 pKa = 10.86ANANVKK140 pKa = 9.73CFPVWQEE147 pKa = 3.1FRR149 pKa = 11.84IPISGTPRR157 pKa = 11.84QKK159 pKa = 10.13MFSTDD164 pKa = 2.14ISTNYY169 pKa = 10.1SVADD173 pKa = 3.64LTRR176 pKa = 11.84TCQGLFAVAIEE187 pKa = 4.24GLTANATAAQTIGRR201 pKa = 11.84CVLEE205 pKa = 4.12TVLQLEE211 pKa = 4.69GLSNPTSS218 pKa = 3.33
Molecular weight: 23.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
726 |
218 |
508 |
363.0 |
41.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.612 ± 1.505 | 2.617 ± 0.39 |
5.096 ± 0.939 | 3.994 ± 0.162 |
4.821 ± 0.802 | 7.3 ± 0.477 |
2.342 ± 0.71 | 5.785 ± 0.089 |
4.959 ± 0.642 | 7.713 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.893 ± 0.298 | 5.51 ± 0.226 |
4.545 ± 0.249 | 4.408 ± 0.139 |
7.713 ± 0.415 | 5.51 ± 0.455 |
5.923 ± 1.391 | 6.887 ± 0.682 |
1.24 ± 0.068 | 4.132 ± 0.687 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
