
Aureimonas sp. Leaf454
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Aurantimonadaceae; Aureimonas; unclassified Aureimonas
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
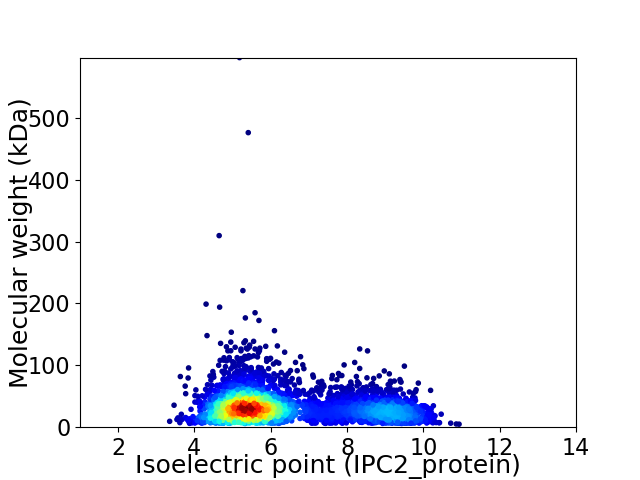
Virtual 2D-PAGE plot for 4218 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
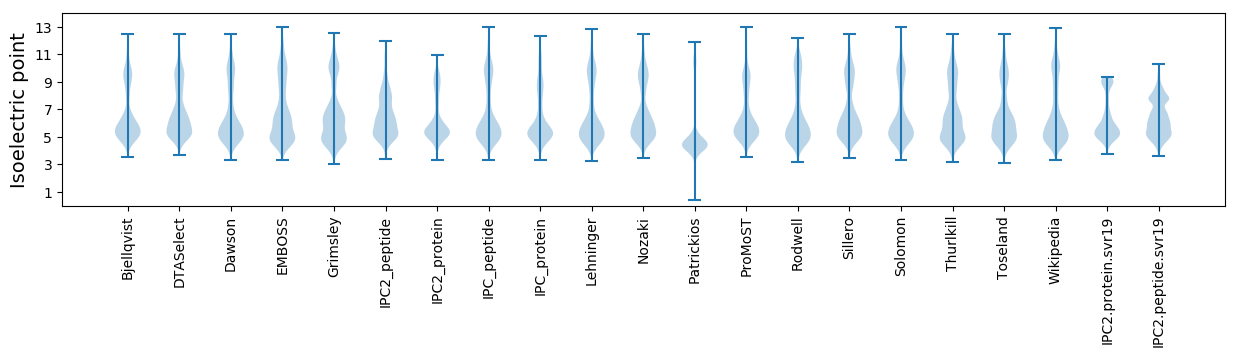
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q6BVB9|A0A0Q6BVB9_9RHIZ Uncharacterized protein OS=Aureimonas sp. Leaf454 OX=1736381 GN=ASG43_13740 PE=3 SV=1
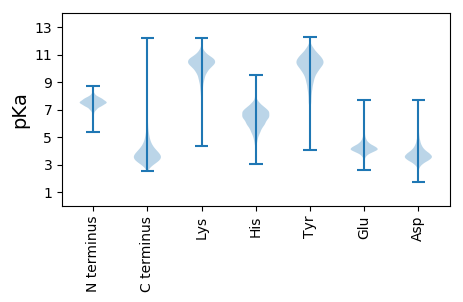
MM1 pKa = 7.36KK2 pKa = 10.37RR3 pKa = 11.84FAILFATTAFVTPAIAADD21 pKa = 3.72VVYY24 pKa = 9.75EE25 pKa = 4.11EE26 pKa = 5.01PPAPVAVMAAPTWTGFYY43 pKa = 10.19IGVQGGGAFNPEE55 pKa = 4.57DD56 pKa = 4.59PDD58 pKa = 3.9SLGIASTFGNATNIGIPNTAGGSLNGAFGGNFASEE93 pKa = 4.12FDD95 pKa = 3.46EE96 pKa = 5.02SFVGGAHH103 pKa = 6.32VGYY106 pKa = 10.06DD107 pKa = 3.45YY108 pKa = 11.86QMDD111 pKa = 3.88NFVLGGVIDD120 pKa = 4.21INALDD125 pKa = 3.86VAQKK129 pKa = 10.5QSAFSNTPAFYY140 pKa = 9.26TAEE143 pKa = 4.08RR144 pKa = 11.84SLDD147 pKa = 3.6FLATARR153 pKa = 11.84LRR155 pKa = 11.84AGYY158 pKa = 10.29AVTPTALAYY167 pKa = 8.63VTGGVAYY174 pKa = 10.44GEE176 pKa = 4.17VDD178 pKa = 3.38YY179 pKa = 11.37SFSSNSPAITAAFPARR195 pKa = 11.84TFEE198 pKa = 4.85DD199 pKa = 3.93EE200 pKa = 5.09DD201 pKa = 4.28DD202 pKa = 3.83FGYY205 pKa = 9.11TVGAGMEE212 pKa = 4.25VMVTEE217 pKa = 4.26NVSFGAEE224 pKa = 3.63YY225 pKa = 10.33LYY227 pKa = 11.03TNLGGDD233 pKa = 3.87SFTTLNGGPFDD244 pKa = 4.6GNGTGLGTGATSFSSNDD261 pKa = 3.2DD262 pKa = 3.53FDD264 pKa = 4.31FHH266 pKa = 6.49TVTAKK271 pKa = 10.46LSYY274 pKa = 10.09RR275 pKa = 11.84FKK277 pKa = 11.26
MM1 pKa = 7.36KK2 pKa = 10.37RR3 pKa = 11.84FAILFATTAFVTPAIAADD21 pKa = 3.72VVYY24 pKa = 9.75EE25 pKa = 4.11EE26 pKa = 5.01PPAPVAVMAAPTWTGFYY43 pKa = 10.19IGVQGGGAFNPEE55 pKa = 4.57DD56 pKa = 4.59PDD58 pKa = 3.9SLGIASTFGNATNIGIPNTAGGSLNGAFGGNFASEE93 pKa = 4.12FDD95 pKa = 3.46EE96 pKa = 5.02SFVGGAHH103 pKa = 6.32VGYY106 pKa = 10.06DD107 pKa = 3.45YY108 pKa = 11.86QMDD111 pKa = 3.88NFVLGGVIDD120 pKa = 4.21INALDD125 pKa = 3.86VAQKK129 pKa = 10.5QSAFSNTPAFYY140 pKa = 9.26TAEE143 pKa = 4.08RR144 pKa = 11.84SLDD147 pKa = 3.6FLATARR153 pKa = 11.84LRR155 pKa = 11.84AGYY158 pKa = 10.29AVTPTALAYY167 pKa = 8.63VTGGVAYY174 pKa = 10.44GEE176 pKa = 4.17VDD178 pKa = 3.38YY179 pKa = 11.37SFSSNSPAITAAFPARR195 pKa = 11.84TFEE198 pKa = 4.85DD199 pKa = 3.93EE200 pKa = 5.09DD201 pKa = 4.28DD202 pKa = 3.83FGYY205 pKa = 9.11TVGAGMEE212 pKa = 4.25VMVTEE217 pKa = 4.26NVSFGAEE224 pKa = 3.63YY225 pKa = 10.33LYY227 pKa = 11.03TNLGGDD233 pKa = 3.87SFTTLNGGPFDD244 pKa = 4.6GNGTGLGTGATSFSSNDD261 pKa = 3.2DD262 pKa = 3.53FDD264 pKa = 4.31FHH266 pKa = 6.49TVTAKK271 pKa = 10.46LSYY274 pKa = 10.09RR275 pKa = 11.84FKK277 pKa = 11.26
Molecular weight: 28.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q6BLI2|A0A0Q6BLI2_9RHIZ Formate--tetrahydrofolate ligase OS=Aureimonas sp. Leaf454 OX=1736381 GN=fhs PE=3 SV=1
MM1 pKa = 7.34SASADD6 pKa = 3.37WTDD9 pKa = 4.93DD10 pKa = 3.21GTTMKK15 pKa = 10.82AIDD18 pKa = 3.82ADD20 pKa = 3.78GFEE23 pKa = 4.84ALFSRR28 pKa = 11.84SIDD31 pKa = 2.99PWNYY35 pKa = 6.7RR36 pKa = 11.84TSRR39 pKa = 11.84FEE41 pKa = 3.48AHH43 pKa = 6.6KK44 pKa = 10.76RR45 pKa = 11.84GVLLKK50 pKa = 10.6ACGLGPYY57 pKa = 10.22GRR59 pKa = 11.84GLEE62 pKa = 4.09LACAIGTTTQALSPLCRR79 pKa = 11.84RR80 pKa = 11.84LLAVDD85 pKa = 4.45ASPTAIEE92 pKa = 3.83EE93 pKa = 4.19ARR95 pKa = 11.84AGRR98 pKa = 11.84YY99 pKa = 7.48AKK101 pKa = 10.23PVTFRR106 pKa = 11.84TARR109 pKa = 11.84LPEE112 pKa = 4.34DD113 pKa = 3.74APRR116 pKa = 11.84GPFDD120 pKa = 5.52LIVVSEE126 pKa = 3.92IAYY129 pKa = 8.86YY130 pKa = 10.48LKK132 pKa = 10.59PRR134 pKa = 11.84ALDD137 pKa = 3.61ALTDD141 pKa = 3.74RR142 pKa = 11.84LLAALAPGGRR152 pKa = 11.84IVVLHH157 pKa = 6.45HH158 pKa = 5.64VTPFADD164 pKa = 3.69AAILPARR171 pKa = 11.84AQAQMTARR179 pKa = 11.84LARR182 pKa = 11.84RR183 pKa = 11.84LAPVLSRR190 pKa = 11.84RR191 pKa = 11.84LGRR194 pKa = 11.84FKK196 pKa = 10.62VAAFEE201 pKa = 3.99RR202 pKa = 11.84RR203 pKa = 11.84RR204 pKa = 11.84RR205 pKa = 3.65
MM1 pKa = 7.34SASADD6 pKa = 3.37WTDD9 pKa = 4.93DD10 pKa = 3.21GTTMKK15 pKa = 10.82AIDD18 pKa = 3.82ADD20 pKa = 3.78GFEE23 pKa = 4.84ALFSRR28 pKa = 11.84SIDD31 pKa = 2.99PWNYY35 pKa = 6.7RR36 pKa = 11.84TSRR39 pKa = 11.84FEE41 pKa = 3.48AHH43 pKa = 6.6KK44 pKa = 10.76RR45 pKa = 11.84GVLLKK50 pKa = 10.6ACGLGPYY57 pKa = 10.22GRR59 pKa = 11.84GLEE62 pKa = 4.09LACAIGTTTQALSPLCRR79 pKa = 11.84RR80 pKa = 11.84LLAVDD85 pKa = 4.45ASPTAIEE92 pKa = 3.83EE93 pKa = 4.19ARR95 pKa = 11.84AGRR98 pKa = 11.84YY99 pKa = 7.48AKK101 pKa = 10.23PVTFRR106 pKa = 11.84TARR109 pKa = 11.84LPEE112 pKa = 4.34DD113 pKa = 3.74APRR116 pKa = 11.84GPFDD120 pKa = 5.52LIVVSEE126 pKa = 3.92IAYY129 pKa = 8.86YY130 pKa = 10.48LKK132 pKa = 10.59PRR134 pKa = 11.84ALDD137 pKa = 3.61ALTDD141 pKa = 3.74RR142 pKa = 11.84LLAALAPGGRR152 pKa = 11.84IVVLHH157 pKa = 6.45HH158 pKa = 5.64VTPFADD164 pKa = 3.69AAILPARR171 pKa = 11.84AQAQMTARR179 pKa = 11.84LARR182 pKa = 11.84RR183 pKa = 11.84LAPVLSRR190 pKa = 11.84RR191 pKa = 11.84LGRR194 pKa = 11.84FKK196 pKa = 10.62VAAFEE201 pKa = 3.99RR202 pKa = 11.84RR203 pKa = 11.84RR204 pKa = 11.84RR205 pKa = 3.65
Molecular weight: 22.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1340498 |
41 |
5673 |
317.8 |
34.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.332 ± 0.061 | 0.73 ± 0.012 |
5.877 ± 0.031 | 5.894 ± 0.036 |
3.69 ± 0.027 | 8.899 ± 0.04 |
1.892 ± 0.02 | 4.995 ± 0.026 |
2.838 ± 0.032 | 10.174 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.321 ± 0.017 | 2.163 ± 0.021 |
5.178 ± 0.03 | 2.652 ± 0.019 |
7.631 ± 0.04 | 5.603 ± 0.029 |
5.368 ± 0.03 | 7.623 ± 0.032 |
1.167 ± 0.015 | 1.973 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
