
Arenibacter aquaticus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Arenibacter
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
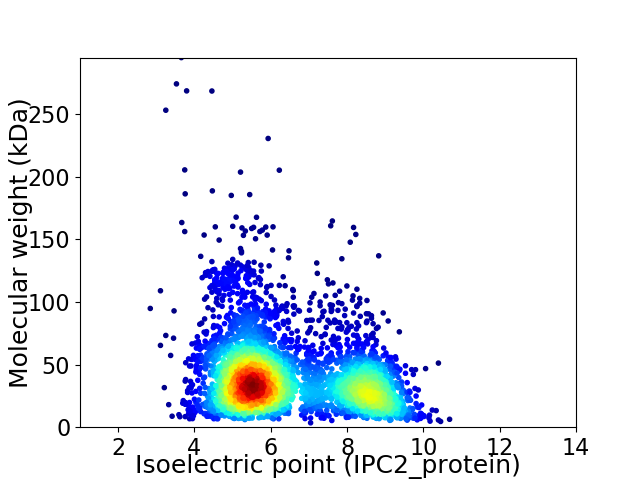
Virtual 2D-PAGE plot for 3975 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S0D2L4|A0A3S0D2L4_9FLAO YjgP/YjgQ family permease OS=Arenibacter aquaticus OX=2489054 GN=EHW67_20215 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 9.52GTVYY6 pKa = 10.2RR7 pKa = 11.84FKK9 pKa = 10.33MLTMASASLCLMASCSSDD27 pKa = 4.51DD28 pKa = 5.6DD29 pKa = 5.41DD30 pKa = 7.2DD31 pKa = 5.54EE32 pKa = 6.05LGNWTDD38 pKa = 3.24KK39 pKa = 11.3SVFDD43 pKa = 3.93GSPRR47 pKa = 11.84SSSAYY52 pKa = 8.55FTIGNIGYY60 pKa = 9.93SGVGYY65 pKa = 10.75DD66 pKa = 3.83GDD68 pKa = 5.23DD69 pKa = 3.56YY70 pKa = 11.37LTSFWAYY77 pKa = 11.09DD78 pKa = 3.24MDD80 pKa = 5.17GDD82 pKa = 4.08YY83 pKa = 10.58WSQKK87 pKa = 10.74ADD89 pKa = 3.71FPGAARR95 pKa = 11.84NAAVGFAIDD104 pKa = 3.44NKK106 pKa = 10.56GYY108 pKa = 10.21IGSGYY113 pKa = 10.56DD114 pKa = 3.77GLDD117 pKa = 3.51EE118 pKa = 5.9LSDD121 pKa = 4.12FYY123 pKa = 11.7SYY125 pKa = 11.4DD126 pKa = 3.39PSSNTWTAIAEE137 pKa = 4.45FPSTPRR143 pKa = 11.84RR144 pKa = 11.84SALAFGINGYY154 pKa = 10.59GYY156 pKa = 10.45FGTGFDD162 pKa = 4.34GDD164 pKa = 3.78NDD166 pKa = 3.98RR167 pKa = 11.84KK168 pKa = 10.91DD169 pKa = 3.13FWKK172 pKa = 10.74YY173 pKa = 11.28DD174 pKa = 3.77PNTDD178 pKa = 2.85SWSEE182 pKa = 3.73LVGFGGDD189 pKa = 3.25KK190 pKa = 10.27RR191 pKa = 11.84RR192 pKa = 11.84SATTFTIGDD201 pKa = 3.93KK202 pKa = 10.92VYY204 pKa = 10.26MGTGVSNGIYY214 pKa = 10.36LDD216 pKa = 4.25DD217 pKa = 3.93FWVFDD222 pKa = 4.03SATEE226 pKa = 3.81SWTQLLDD233 pKa = 3.75LDD235 pKa = 4.83EE236 pKa = 5.2EE237 pKa = 4.87DD238 pKa = 4.87DD239 pKa = 4.03YY240 pKa = 12.08SIVRR244 pKa = 11.84SNAVGFTLNDD254 pKa = 3.15YY255 pKa = 11.12GYY257 pKa = 9.93IATGTSGGTLQSVWEE272 pKa = 4.25YY273 pKa = 11.53DD274 pKa = 3.51PSNDD278 pKa = 2.54TWEE281 pKa = 4.45AKK283 pKa = 9.7TSFEE287 pKa = 3.95RR288 pKa = 11.84TSRR291 pKa = 11.84QDD293 pKa = 3.15AVVFSNNVRR302 pKa = 11.84AFIGLGRR309 pKa = 11.84TGSLYY314 pKa = 10.87LDD316 pKa = 4.28DD317 pKa = 5.7LEE319 pKa = 5.55EE320 pKa = 4.09FFPYY324 pKa = 10.34DD325 pKa = 3.6EE326 pKa = 5.37YY327 pKa = 11.74DD328 pKa = 4.78DD329 pKa = 4.65EE330 pKa = 5.01DD331 pKa = 3.64
MM1 pKa = 7.63KK2 pKa = 9.52GTVYY6 pKa = 10.2RR7 pKa = 11.84FKK9 pKa = 10.33MLTMASASLCLMASCSSDD27 pKa = 4.51DD28 pKa = 5.6DD29 pKa = 5.41DD30 pKa = 7.2DD31 pKa = 5.54EE32 pKa = 6.05LGNWTDD38 pKa = 3.24KK39 pKa = 11.3SVFDD43 pKa = 3.93GSPRR47 pKa = 11.84SSSAYY52 pKa = 8.55FTIGNIGYY60 pKa = 9.93SGVGYY65 pKa = 10.75DD66 pKa = 3.83GDD68 pKa = 5.23DD69 pKa = 3.56YY70 pKa = 11.37LTSFWAYY77 pKa = 11.09DD78 pKa = 3.24MDD80 pKa = 5.17GDD82 pKa = 4.08YY83 pKa = 10.58WSQKK87 pKa = 10.74ADD89 pKa = 3.71FPGAARR95 pKa = 11.84NAAVGFAIDD104 pKa = 3.44NKK106 pKa = 10.56GYY108 pKa = 10.21IGSGYY113 pKa = 10.56DD114 pKa = 3.77GLDD117 pKa = 3.51EE118 pKa = 5.9LSDD121 pKa = 4.12FYY123 pKa = 11.7SYY125 pKa = 11.4DD126 pKa = 3.39PSSNTWTAIAEE137 pKa = 4.45FPSTPRR143 pKa = 11.84RR144 pKa = 11.84SALAFGINGYY154 pKa = 10.59GYY156 pKa = 10.45FGTGFDD162 pKa = 4.34GDD164 pKa = 3.78NDD166 pKa = 3.98RR167 pKa = 11.84KK168 pKa = 10.91DD169 pKa = 3.13FWKK172 pKa = 10.74YY173 pKa = 11.28DD174 pKa = 3.77PNTDD178 pKa = 2.85SWSEE182 pKa = 3.73LVGFGGDD189 pKa = 3.25KK190 pKa = 10.27RR191 pKa = 11.84RR192 pKa = 11.84SATTFTIGDD201 pKa = 3.93KK202 pKa = 10.92VYY204 pKa = 10.26MGTGVSNGIYY214 pKa = 10.36LDD216 pKa = 4.25DD217 pKa = 3.93FWVFDD222 pKa = 4.03SATEE226 pKa = 3.81SWTQLLDD233 pKa = 3.75LDD235 pKa = 4.83EE236 pKa = 5.2EE237 pKa = 4.87DD238 pKa = 4.87DD239 pKa = 4.03YY240 pKa = 12.08SIVRR244 pKa = 11.84SNAVGFTLNDD254 pKa = 3.15YY255 pKa = 11.12GYY257 pKa = 9.93IATGTSGGTLQSVWEE272 pKa = 4.25YY273 pKa = 11.53DD274 pKa = 3.51PSNDD278 pKa = 2.54TWEE281 pKa = 4.45AKK283 pKa = 9.7TSFEE287 pKa = 3.95RR288 pKa = 11.84TSRR291 pKa = 11.84QDD293 pKa = 3.15AVVFSNNVRR302 pKa = 11.84AFIGLGRR309 pKa = 11.84TGSLYY314 pKa = 10.87LDD316 pKa = 4.28DD317 pKa = 5.7LEE319 pKa = 5.55EE320 pKa = 4.09FFPYY324 pKa = 10.34DD325 pKa = 3.6EE326 pKa = 5.37YY327 pKa = 11.74DD328 pKa = 4.78DD329 pKa = 4.65EE330 pKa = 5.01DD331 pKa = 3.64
Molecular weight: 36.81 kDa
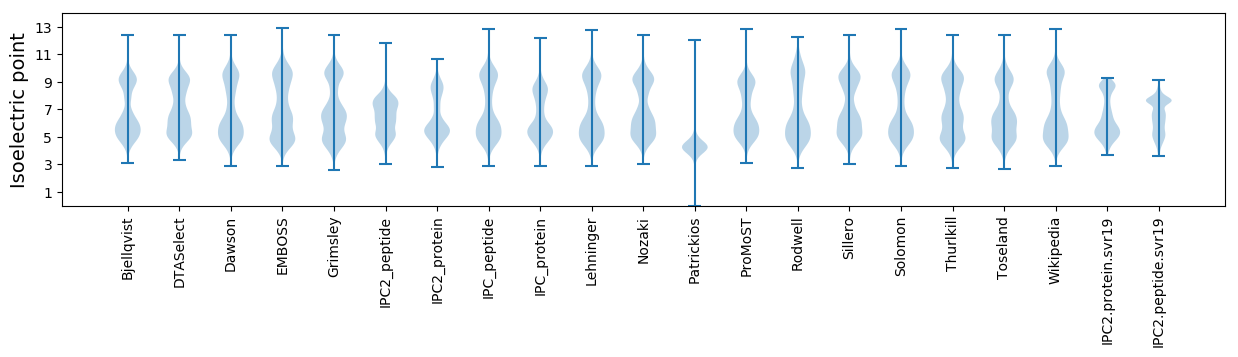
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S0BVA2|A0A3S0BVA2_9FLAO Response regulator OS=Arenibacter aquaticus OX=2489054 GN=EHW67_19735 PE=4 SV=1
MM1 pKa = 7.79RR2 pKa = 11.84SFRR5 pKa = 11.84FIDD8 pKa = 3.26QEE10 pKa = 3.98FRR12 pKa = 11.84DD13 pKa = 3.94VSTLLRR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 8.11TKK23 pKa = 9.89IHH25 pKa = 5.95FKK27 pKa = 10.2KK28 pKa = 10.42DD29 pKa = 3.35YY30 pKa = 8.87MLLRR34 pKa = 11.84SIPSIGPIVASGILSEE50 pKa = 5.23LGDD53 pKa = 3.65LRR55 pKa = 11.84RR56 pKa = 11.84FKK58 pKa = 10.69NIKK61 pKa = 9.53HH62 pKa = 5.54LAGYY66 pKa = 9.8VGLAPGIYY74 pKa = 9.76QSGDD78 pKa = 3.37TVRR81 pKa = 11.84HH82 pKa = 5.63TGVSMRR88 pKa = 11.84AHH90 pKa = 6.49RR91 pKa = 11.84LIRR94 pKa = 11.84SYY96 pKa = 10.66FIEE99 pKa = 4.74ASWQAIRR106 pKa = 11.84TDD108 pKa = 3.41PVMQAFYY115 pKa = 10.63RR116 pKa = 11.84KK117 pKa = 8.99HH118 pKa = 5.74HH119 pKa = 5.88GKK121 pKa = 9.2NVKK124 pKa = 10.36SIIVKK129 pKa = 8.57VARR132 pKa = 11.84KK133 pKa = 9.33LLSRR137 pKa = 11.84TLAVIKK143 pKa = 10.07TEE145 pKa = 3.95TPYY148 pKa = 11.19VIGVLEE154 pKa = 3.97
MM1 pKa = 7.79RR2 pKa = 11.84SFRR5 pKa = 11.84FIDD8 pKa = 3.26QEE10 pKa = 3.98FRR12 pKa = 11.84DD13 pKa = 3.94VSTLLRR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 8.11TKK23 pKa = 9.89IHH25 pKa = 5.95FKK27 pKa = 10.2KK28 pKa = 10.42DD29 pKa = 3.35YY30 pKa = 8.87MLLRR34 pKa = 11.84SIPSIGPIVASGILSEE50 pKa = 5.23LGDD53 pKa = 3.65LRR55 pKa = 11.84RR56 pKa = 11.84FKK58 pKa = 10.69NIKK61 pKa = 9.53HH62 pKa = 5.54LAGYY66 pKa = 9.8VGLAPGIYY74 pKa = 9.76QSGDD78 pKa = 3.37TVRR81 pKa = 11.84HH82 pKa = 5.63TGVSMRR88 pKa = 11.84AHH90 pKa = 6.49RR91 pKa = 11.84LIRR94 pKa = 11.84SYY96 pKa = 10.66FIEE99 pKa = 4.74ASWQAIRR106 pKa = 11.84TDD108 pKa = 3.41PVMQAFYY115 pKa = 10.63RR116 pKa = 11.84KK117 pKa = 8.99HH118 pKa = 5.74HH119 pKa = 5.88GKK121 pKa = 9.2NVKK124 pKa = 10.36SIIVKK129 pKa = 8.57VARR132 pKa = 11.84KK133 pKa = 9.33LLSRR137 pKa = 11.84TLAVIKK143 pKa = 10.07TEE145 pKa = 3.95TPYY148 pKa = 11.19VIGVLEE154 pKa = 3.97
Molecular weight: 17.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1516357 |
29 |
2814 |
381.5 |
42.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.539 ± 0.034 | 0.725 ± 0.012 |
5.741 ± 0.035 | 6.528 ± 0.036 |
4.84 ± 0.025 | 7.143 ± 0.054 |
1.878 ± 0.018 | 7.213 ± 0.035 |
7.306 ± 0.042 | 9.371 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.333 ± 0.015 | 5.867 ± 0.035 |
3.759 ± 0.022 | 3.416 ± 0.022 |
3.667 ± 0.023 | 6.476 ± 0.028 |
5.587 ± 0.04 | 6.234 ± 0.029 |
1.251 ± 0.017 | 4.127 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
