
Spinach cryptic virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
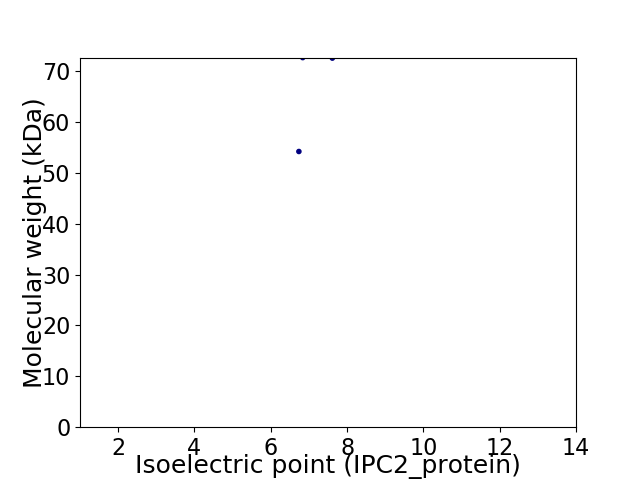
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1P8NQ96|A0A1P8NQ96_9VIRU RNA-dependent RNA polymerase OS=Spinach cryptic virus 1 OX=1937817 GN=RdRp PE=4 SV=1
MM1 pKa = 7.49NPEE4 pKa = 4.14TPLADD9 pKa = 4.09PNGPTPPHH17 pKa = 6.31GAAAQPTPGQHH28 pKa = 6.07IPAGTPSTGAVHH40 pKa = 6.59LTPAPQPKK48 pKa = 9.75RR49 pKa = 11.84NRR51 pKa = 11.84APRR54 pKa = 11.84GPFDD58 pKa = 3.76NASSTSAPGAPALLEE73 pKa = 3.95LAAAYY78 pKa = 9.21PMSTEE83 pKa = 3.91QRR85 pKa = 11.84RR86 pKa = 11.84SPNTFVPDD94 pKa = 3.45AQMLFHH100 pKa = 6.92VLSLCDD106 pKa = 4.12NMMLSTDD113 pKa = 3.42RR114 pKa = 11.84FTRR117 pKa = 11.84SAPAWLPIVSQLYY130 pKa = 10.17ISILWFTMILKK141 pKa = 10.32VYY143 pKa = 10.14VNSSYY148 pKa = 11.54GAMHH152 pKa = 6.98AHH154 pKa = 7.6LLNILVAHH162 pKa = 6.31LRR164 pKa = 11.84IDD166 pKa = 3.39EE167 pKa = 4.21CLIPGPLVPFFQSLGAINGPFEE189 pKa = 4.86WIGDD193 pKa = 3.76IVPIIPEE200 pKa = 4.79FSQLWNTTSFYY211 pKa = 9.92PTDD214 pKa = 3.18SFARR218 pKa = 11.84IFPIPAILLDD228 pKa = 3.6QLHH231 pKa = 6.19YY232 pKa = 9.92FASITVTEE240 pKa = 4.02QEE242 pKa = 4.4TIFGHH247 pKa = 5.62FEE249 pKa = 3.51WYY251 pKa = 10.4RR252 pKa = 11.84NIFSLGQGTFNPRR265 pKa = 11.84LRR267 pKa = 11.84LGPQLCGSLYY277 pKa = 9.56ATEE280 pKa = 4.24SQVTNARR287 pKa = 11.84TFWSAALRR295 pKa = 11.84TGITRR300 pKa = 11.84TNAAANQPLFSSYY313 pKa = 10.19PQLLGFVDD321 pKa = 4.34QNDD324 pKa = 3.29AMQLDD329 pKa = 5.02WFQHH333 pKa = 4.56TAIVMQKK340 pKa = 8.62YY341 pKa = 8.82CQYY344 pKa = 10.77FNGSVPLKK352 pKa = 10.26SISPTGIGAVAVHH365 pKa = 6.93GYY367 pKa = 9.75PNYY370 pKa = 10.71SGAARR375 pKa = 11.84TWMYY379 pKa = 10.17PATNTLSSFKK389 pKa = 10.48SSRR392 pKa = 11.84FSALRR397 pKa = 11.84EE398 pKa = 3.96IPALLTVRR406 pKa = 11.84FEE408 pKa = 4.42HH409 pKa = 7.19ADD411 pKa = 3.49HH412 pKa = 6.48EE413 pKa = 4.53LEE415 pKa = 4.27EE416 pKa = 4.19QAEE419 pKa = 4.35QYY421 pKa = 11.19AIATHH426 pKa = 6.91TNMCWSKK433 pKa = 11.38NLTTQNNWTQVAEE446 pKa = 4.35ANVHH450 pKa = 6.18SGTYY454 pKa = 9.53WSMTPHH460 pKa = 7.14RR461 pKa = 11.84AHH463 pKa = 6.79ASVRR467 pKa = 11.84LKK469 pKa = 9.2TQYY472 pKa = 10.88AQLVASRR479 pKa = 11.84YY480 pKa = 8.33HH481 pKa = 4.53QQAANRR487 pKa = 11.84VV488 pKa = 3.59
MM1 pKa = 7.49NPEE4 pKa = 4.14TPLADD9 pKa = 4.09PNGPTPPHH17 pKa = 6.31GAAAQPTPGQHH28 pKa = 6.07IPAGTPSTGAVHH40 pKa = 6.59LTPAPQPKK48 pKa = 9.75RR49 pKa = 11.84NRR51 pKa = 11.84APRR54 pKa = 11.84GPFDD58 pKa = 3.76NASSTSAPGAPALLEE73 pKa = 3.95LAAAYY78 pKa = 9.21PMSTEE83 pKa = 3.91QRR85 pKa = 11.84RR86 pKa = 11.84SPNTFVPDD94 pKa = 3.45AQMLFHH100 pKa = 6.92VLSLCDD106 pKa = 4.12NMMLSTDD113 pKa = 3.42RR114 pKa = 11.84FTRR117 pKa = 11.84SAPAWLPIVSQLYY130 pKa = 10.17ISILWFTMILKK141 pKa = 10.32VYY143 pKa = 10.14VNSSYY148 pKa = 11.54GAMHH152 pKa = 6.98AHH154 pKa = 7.6LLNILVAHH162 pKa = 6.31LRR164 pKa = 11.84IDD166 pKa = 3.39EE167 pKa = 4.21CLIPGPLVPFFQSLGAINGPFEE189 pKa = 4.86WIGDD193 pKa = 3.76IVPIIPEE200 pKa = 4.79FSQLWNTTSFYY211 pKa = 9.92PTDD214 pKa = 3.18SFARR218 pKa = 11.84IFPIPAILLDD228 pKa = 3.6QLHH231 pKa = 6.19YY232 pKa = 9.92FASITVTEE240 pKa = 4.02QEE242 pKa = 4.4TIFGHH247 pKa = 5.62FEE249 pKa = 3.51WYY251 pKa = 10.4RR252 pKa = 11.84NIFSLGQGTFNPRR265 pKa = 11.84LRR267 pKa = 11.84LGPQLCGSLYY277 pKa = 9.56ATEE280 pKa = 4.24SQVTNARR287 pKa = 11.84TFWSAALRR295 pKa = 11.84TGITRR300 pKa = 11.84TNAAANQPLFSSYY313 pKa = 10.19PQLLGFVDD321 pKa = 4.34QNDD324 pKa = 3.29AMQLDD329 pKa = 5.02WFQHH333 pKa = 4.56TAIVMQKK340 pKa = 8.62YY341 pKa = 8.82CQYY344 pKa = 10.77FNGSVPLKK352 pKa = 10.26SISPTGIGAVAVHH365 pKa = 6.93GYY367 pKa = 9.75PNYY370 pKa = 10.71SGAARR375 pKa = 11.84TWMYY379 pKa = 10.17PATNTLSSFKK389 pKa = 10.48SSRR392 pKa = 11.84FSALRR397 pKa = 11.84EE398 pKa = 3.96IPALLTVRR406 pKa = 11.84FEE408 pKa = 4.42HH409 pKa = 7.19ADD411 pKa = 3.49HH412 pKa = 6.48EE413 pKa = 4.53LEE415 pKa = 4.27EE416 pKa = 4.19QAEE419 pKa = 4.35QYY421 pKa = 11.19AIATHH426 pKa = 6.91TNMCWSKK433 pKa = 11.38NLTTQNNWTQVAEE446 pKa = 4.35ANVHH450 pKa = 6.18SGTYY454 pKa = 9.53WSMTPHH460 pKa = 7.14RR461 pKa = 11.84AHH463 pKa = 6.79ASVRR467 pKa = 11.84LKK469 pKa = 9.2TQYY472 pKa = 10.88AQLVASRR479 pKa = 11.84YY480 pKa = 8.33HH481 pKa = 4.53QQAANRR487 pKa = 11.84VV488 pKa = 3.59
Molecular weight: 54.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8NQ96|A0A1P8NQ96_9VIRU RNA-dependent RNA polymerase OS=Spinach cryptic virus 1 OX=1937817 GN=RdRp PE=4 SV=1
MM1 pKa = 8.32DD2 pKa = 4.54YY3 pKa = 9.87LTSAFNRR10 pKa = 11.84ITHH13 pKa = 7.05WFTKK17 pKa = 8.13PTNLEE22 pKa = 4.11YY23 pKa = 10.41IGDD26 pKa = 4.0YY27 pKa = 8.46STPPSIIRR35 pKa = 11.84VNEE38 pKa = 3.64VAIANHH44 pKa = 6.41KK45 pKa = 8.46KK46 pKa = 6.56TLRR49 pKa = 11.84YY50 pKa = 9.94AFEE53 pKa = 4.03QYY55 pKa = 10.84LYY57 pKa = 9.86DD58 pKa = 3.33HH59 pKa = 8.11EE60 pKa = 4.34YY61 pKa = 11.34RR62 pKa = 11.84MIVDD66 pKa = 4.18EE67 pKa = 4.34YY68 pKa = 10.64RR69 pKa = 11.84RR70 pKa = 11.84SDD72 pKa = 3.1IDD74 pKa = 3.44QEE76 pKa = 4.71SILADD81 pKa = 4.36FFSGDD86 pKa = 3.85IEE88 pKa = 4.44PFDD91 pKa = 3.74IPFDD95 pKa = 3.52EE96 pKa = 4.69HH97 pKa = 8.08VEE99 pKa = 3.99HH100 pKa = 7.35GLRR103 pKa = 11.84CMANAFRR110 pKa = 11.84PPRR113 pKa = 11.84PCRR116 pKa = 11.84PAHH119 pKa = 6.19ILDD122 pKa = 3.78VKK124 pKa = 10.61HH125 pKa = 6.44GYY127 pKa = 8.11PYY129 pKa = 10.43KK130 pKa = 10.06WNVNAEE136 pKa = 4.45PPFSTDD142 pKa = 3.47PYY144 pKa = 10.61FLSQRR149 pKa = 11.84KK150 pKa = 6.53TFGEE154 pKa = 4.98FIQMHH159 pKa = 6.4EE160 pKa = 4.3YY161 pKa = 10.66EE162 pKa = 5.81HH163 pKa = 6.97IDD165 pKa = 3.09KK166 pKa = 11.16ADD168 pKa = 3.68FFRR171 pKa = 11.84RR172 pKa = 11.84HH173 pKa = 6.43PNTEE177 pKa = 3.52SHH179 pKa = 7.34DD180 pKa = 3.85FLTTVVPPKK189 pKa = 10.49FGYY192 pKa = 9.69LKK194 pKa = 11.1SMVFSWTRR202 pKa = 11.84RR203 pKa = 11.84WHH205 pKa = 6.16HH206 pKa = 5.75VIKK209 pKa = 9.97TGFQDD214 pKa = 3.5LTGLEE219 pKa = 4.16NSGYY223 pKa = 10.32LYY225 pKa = 10.98NRR227 pKa = 11.84FIFPMLLHH235 pKa = 6.66TKK237 pKa = 8.3TAIVKK242 pKa = 10.57KK243 pKa = 10.26NDD245 pKa = 3.58PNKK248 pKa = 9.91MRR250 pKa = 11.84TIWGAPKK257 pKa = 9.48PWIIAEE263 pKa = 4.17TMLYY267 pKa = 9.2WEE269 pKa = 4.49YY270 pKa = 10.8IAWIKK275 pKa = 10.52LNSGKK280 pKa = 8.34TPMLWGFEE288 pKa = 4.25TFTGGWFRR296 pKa = 11.84LNNILFCGLIKK307 pKa = 10.6QSFLTLDD314 pKa = 3.5WSRR317 pKa = 11.84FDD319 pKa = 3.08KK320 pKa = 10.75RR321 pKa = 11.84AYY323 pKa = 10.1FSLLRR328 pKa = 11.84RR329 pKa = 11.84ILYY332 pKa = 6.74TARR335 pKa = 11.84TFLTFDD341 pKa = 2.91EE342 pKa = 6.09GYY344 pKa = 10.31VPTHH348 pKa = 7.06AAPTHH353 pKa = 5.97PKK355 pKa = 9.28WNHH358 pKa = 3.88QKK360 pKa = 11.04ALNLEE365 pKa = 4.43RR366 pKa = 11.84LWLWTLEE373 pKa = 4.18NLFEE377 pKa = 4.94APIILPDD384 pKa = 2.89GRR386 pKa = 11.84MYY388 pKa = 10.65RR389 pKa = 11.84RR390 pKa = 11.84HH391 pKa = 5.78FAGIPSGLFITQLLDD406 pKa = 2.7SWYY409 pKa = 9.85NYY411 pKa = 9.33TMLATLLSALGFDD424 pKa = 4.3PKK426 pKa = 11.16YY427 pKa = 10.85CIIKK431 pKa = 9.83VQGDD435 pKa = 3.45DD436 pKa = 4.79SIIKK440 pKa = 8.74LTTLIPRR447 pKa = 11.84DD448 pKa = 3.31QHH450 pKa = 6.01EE451 pKa = 4.46NFMVQLTALATTYY464 pKa = 10.69FNATINVKK472 pKa = 10.27KK473 pKa = 10.86SEE475 pKa = 4.06IRR477 pKa = 11.84NTLNGCEE484 pKa = 3.86VLSYY488 pKa = 10.75KK489 pKa = 10.05NHH491 pKa = 6.42NGFPHH496 pKa = 6.91RR497 pKa = 11.84DD498 pKa = 4.01EE499 pKa = 4.17ITMLAQFYY507 pKa = 7.85HH508 pKa = 6.12TKK510 pKa = 10.61ARR512 pKa = 11.84DD513 pKa = 3.58PTPEE517 pKa = 3.33ITMAQAIGFAYY528 pKa = 10.22ASCGNNKK535 pKa = 9.26RR536 pKa = 11.84VLWILRR542 pKa = 11.84DD543 pKa = 3.15IYY545 pKa = 11.37NFYY548 pKa = 11.2KK549 pKa = 10.24MRR551 pKa = 11.84DD552 pKa = 3.55VTPNRR557 pKa = 11.84AGLTLTFGDD566 pKa = 4.2SPDD569 pKa = 3.21VFMPEE574 pKa = 3.47IPLDD578 pKa = 3.71HH579 pKa = 7.05FPTITEE585 pKa = 3.31IRR587 pKa = 11.84RR588 pKa = 11.84YY589 pKa = 7.58LTCSEE594 pKa = 4.24YY595 pKa = 10.89RR596 pKa = 11.84NEE598 pKa = 4.15AQDD601 pKa = 3.1ARR603 pKa = 11.84TWPRR607 pKa = 11.84SLFISGPAEE616 pKa = 3.67
MM1 pKa = 8.32DD2 pKa = 4.54YY3 pKa = 9.87LTSAFNRR10 pKa = 11.84ITHH13 pKa = 7.05WFTKK17 pKa = 8.13PTNLEE22 pKa = 4.11YY23 pKa = 10.41IGDD26 pKa = 4.0YY27 pKa = 8.46STPPSIIRR35 pKa = 11.84VNEE38 pKa = 3.64VAIANHH44 pKa = 6.41KK45 pKa = 8.46KK46 pKa = 6.56TLRR49 pKa = 11.84YY50 pKa = 9.94AFEE53 pKa = 4.03QYY55 pKa = 10.84LYY57 pKa = 9.86DD58 pKa = 3.33HH59 pKa = 8.11EE60 pKa = 4.34YY61 pKa = 11.34RR62 pKa = 11.84MIVDD66 pKa = 4.18EE67 pKa = 4.34YY68 pKa = 10.64RR69 pKa = 11.84RR70 pKa = 11.84SDD72 pKa = 3.1IDD74 pKa = 3.44QEE76 pKa = 4.71SILADD81 pKa = 4.36FFSGDD86 pKa = 3.85IEE88 pKa = 4.44PFDD91 pKa = 3.74IPFDD95 pKa = 3.52EE96 pKa = 4.69HH97 pKa = 8.08VEE99 pKa = 3.99HH100 pKa = 7.35GLRR103 pKa = 11.84CMANAFRR110 pKa = 11.84PPRR113 pKa = 11.84PCRR116 pKa = 11.84PAHH119 pKa = 6.19ILDD122 pKa = 3.78VKK124 pKa = 10.61HH125 pKa = 6.44GYY127 pKa = 8.11PYY129 pKa = 10.43KK130 pKa = 10.06WNVNAEE136 pKa = 4.45PPFSTDD142 pKa = 3.47PYY144 pKa = 10.61FLSQRR149 pKa = 11.84KK150 pKa = 6.53TFGEE154 pKa = 4.98FIQMHH159 pKa = 6.4EE160 pKa = 4.3YY161 pKa = 10.66EE162 pKa = 5.81HH163 pKa = 6.97IDD165 pKa = 3.09KK166 pKa = 11.16ADD168 pKa = 3.68FFRR171 pKa = 11.84RR172 pKa = 11.84HH173 pKa = 6.43PNTEE177 pKa = 3.52SHH179 pKa = 7.34DD180 pKa = 3.85FLTTVVPPKK189 pKa = 10.49FGYY192 pKa = 9.69LKK194 pKa = 11.1SMVFSWTRR202 pKa = 11.84RR203 pKa = 11.84WHH205 pKa = 6.16HH206 pKa = 5.75VIKK209 pKa = 9.97TGFQDD214 pKa = 3.5LTGLEE219 pKa = 4.16NSGYY223 pKa = 10.32LYY225 pKa = 10.98NRR227 pKa = 11.84FIFPMLLHH235 pKa = 6.66TKK237 pKa = 8.3TAIVKK242 pKa = 10.57KK243 pKa = 10.26NDD245 pKa = 3.58PNKK248 pKa = 9.91MRR250 pKa = 11.84TIWGAPKK257 pKa = 9.48PWIIAEE263 pKa = 4.17TMLYY267 pKa = 9.2WEE269 pKa = 4.49YY270 pKa = 10.8IAWIKK275 pKa = 10.52LNSGKK280 pKa = 8.34TPMLWGFEE288 pKa = 4.25TFTGGWFRR296 pKa = 11.84LNNILFCGLIKK307 pKa = 10.6QSFLTLDD314 pKa = 3.5WSRR317 pKa = 11.84FDD319 pKa = 3.08KK320 pKa = 10.75RR321 pKa = 11.84AYY323 pKa = 10.1FSLLRR328 pKa = 11.84RR329 pKa = 11.84ILYY332 pKa = 6.74TARR335 pKa = 11.84TFLTFDD341 pKa = 2.91EE342 pKa = 6.09GYY344 pKa = 10.31VPTHH348 pKa = 7.06AAPTHH353 pKa = 5.97PKK355 pKa = 9.28WNHH358 pKa = 3.88QKK360 pKa = 11.04ALNLEE365 pKa = 4.43RR366 pKa = 11.84LWLWTLEE373 pKa = 4.18NLFEE377 pKa = 4.94APIILPDD384 pKa = 2.89GRR386 pKa = 11.84MYY388 pKa = 10.65RR389 pKa = 11.84RR390 pKa = 11.84HH391 pKa = 5.78FAGIPSGLFITQLLDD406 pKa = 2.7SWYY409 pKa = 9.85NYY411 pKa = 9.33TMLATLLSALGFDD424 pKa = 4.3PKK426 pKa = 11.16YY427 pKa = 10.85CIIKK431 pKa = 9.83VQGDD435 pKa = 3.45DD436 pKa = 4.79SIIKK440 pKa = 8.74LTTLIPRR447 pKa = 11.84DD448 pKa = 3.31QHH450 pKa = 6.01EE451 pKa = 4.46NFMVQLTALATTYY464 pKa = 10.69FNATINVKK472 pKa = 10.27KK473 pKa = 10.86SEE475 pKa = 4.06IRR477 pKa = 11.84NTLNGCEE484 pKa = 3.86VLSYY488 pKa = 10.75KK489 pKa = 10.05NHH491 pKa = 6.42NGFPHH496 pKa = 6.91RR497 pKa = 11.84DD498 pKa = 4.01EE499 pKa = 4.17ITMLAQFYY507 pKa = 7.85HH508 pKa = 6.12TKK510 pKa = 10.61ARR512 pKa = 11.84DD513 pKa = 3.58PTPEE517 pKa = 3.33ITMAQAIGFAYY528 pKa = 10.22ASCGNNKK535 pKa = 9.26RR536 pKa = 11.84VLWILRR542 pKa = 11.84DD543 pKa = 3.15IYY545 pKa = 11.37NFYY548 pKa = 11.2KK549 pKa = 10.24MRR551 pKa = 11.84DD552 pKa = 3.55VTPNRR557 pKa = 11.84AGLTLTFGDD566 pKa = 4.2SPDD569 pKa = 3.21VFMPEE574 pKa = 3.47IPLDD578 pKa = 3.71HH579 pKa = 7.05FPTITEE585 pKa = 3.31IRR587 pKa = 11.84RR588 pKa = 11.84YY589 pKa = 7.58LTCSEE594 pKa = 4.24YY595 pKa = 10.89RR596 pKa = 11.84NEE598 pKa = 4.15AQDD601 pKa = 3.1ARR603 pKa = 11.84TWPRR607 pKa = 11.84SLFISGPAEE616 pKa = 3.67
Molecular weight: 72.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1104 |
488 |
616 |
552.0 |
63.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.062 ± 1.785 | 1.087 ± 0.04 |
4.257 ± 1.016 | 4.438 ± 0.609 |
6.159 ± 0.661 | 4.801 ± 0.205 |
3.714 ± 0.016 | 6.25 ± 0.719 |
3.351 ± 1.222 | 8.877 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.536 ± 0.049 | 5.072 ± 0.032 |
7.246 ± 0.737 | 3.804 ± 1.233 |
5.707 ± 0.633 | 5.888 ± 1.211 |
7.971 ± 0.013 | 3.714 ± 0.507 |
2.536 ± 0.18 | 4.529 ± 0.536 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
