
Ustilaginoidea virens partitivirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
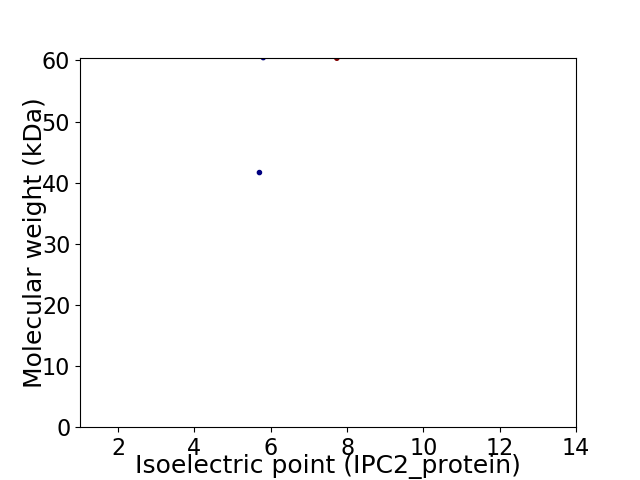
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5M8T3|S5M8T3_9VIRU Uncharacterized protein OS=Ustilaginoidea virens partitivirus 2 OX=1415665 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 10.65NEE4 pKa = 3.98IAFQPGKK11 pKa = 9.65FSPVIEE17 pKa = 4.17VSQRR21 pKa = 11.84QSGVPAPADD30 pKa = 3.37TSFDD34 pKa = 3.48GLEE37 pKa = 4.1EE38 pKa = 4.19VALTTLQVATSSARR52 pKa = 11.84GSGAVIQDD60 pKa = 3.71SVPLSKK66 pKa = 10.87LSMVIGYY73 pKa = 9.11LLAARR78 pKa = 11.84NIAEE82 pKa = 3.96QLLFIGVSTRR92 pKa = 11.84MSTPLRR98 pKa = 11.84APNGNFLEE106 pKa = 5.13PIVRR110 pKa = 11.84AYY112 pKa = 11.01NFYY115 pKa = 11.14GHH117 pKa = 6.84FEE119 pKa = 4.33HH120 pKa = 7.35EE121 pKa = 4.6SKK123 pKa = 9.93TFVTRR128 pKa = 11.84GLEE131 pKa = 4.09TLFIRR136 pKa = 11.84YY137 pKa = 8.79LFGLRR142 pKa = 11.84QFATTGVSDD151 pKa = 4.61GSAHH155 pKa = 5.0TVRR158 pKa = 11.84VLSNQNHH165 pKa = 5.62NFSDD169 pKa = 3.5IDD171 pKa = 3.86LDD173 pKa = 4.46FYY175 pKa = 11.55YY176 pKa = 10.64LSEE179 pKa = 5.01AGEE182 pKa = 3.99IAFGKK187 pKa = 10.1RR188 pKa = 11.84YY189 pKa = 8.78PLKK192 pKa = 10.01EE193 pKa = 3.95YY194 pKa = 10.48LIHH197 pKa = 5.8LVKK200 pKa = 10.64YY201 pKa = 10.42ASDD204 pKa = 3.57FGDD207 pKa = 3.17VDD209 pKa = 3.4EE210 pKa = 6.36RR211 pKa = 11.84IPLLQRR217 pKa = 11.84ILDD220 pKa = 3.76VTTEE224 pKa = 4.11DD225 pKa = 4.42GLNQFLRR232 pKa = 11.84YY233 pKa = 9.57ARR235 pKa = 11.84RR236 pKa = 11.84LPDD239 pKa = 2.97WQTPPRR245 pKa = 11.84EE246 pKa = 4.16FTPDD250 pKa = 3.22PSDD253 pKa = 3.21KK254 pKa = 10.59HH255 pKa = 6.76KK256 pKa = 11.32SAMKK260 pKa = 9.87EE261 pKa = 3.92LFGAAYY267 pKa = 8.26STAGQSIPVSKK278 pKa = 9.42FTEE281 pKa = 4.71SINLAYY287 pKa = 10.45ALLRR291 pKa = 11.84RR292 pKa = 11.84VSLEE296 pKa = 3.48RR297 pKa = 11.84QYY299 pKa = 12.08GMKK302 pKa = 9.41TVQIPRR308 pKa = 11.84YY309 pKa = 8.61EE310 pKa = 4.43GGSPSQMASYY320 pKa = 10.69QDD322 pKa = 4.16DD323 pKa = 4.1VLFSTIPLSLADD335 pKa = 3.38STAAVGFTTSYY346 pKa = 11.25GSLARR351 pKa = 11.84YY352 pKa = 7.09TSAPGFEE359 pKa = 4.16KK360 pKa = 10.91DD361 pKa = 3.32EE362 pKa = 4.27LVRR365 pKa = 11.84EE366 pKa = 4.33LVSQSLVLRR375 pKa = 4.52
MM1 pKa = 7.36KK2 pKa = 10.65NEE4 pKa = 3.98IAFQPGKK11 pKa = 9.65FSPVIEE17 pKa = 4.17VSQRR21 pKa = 11.84QSGVPAPADD30 pKa = 3.37TSFDD34 pKa = 3.48GLEE37 pKa = 4.1EE38 pKa = 4.19VALTTLQVATSSARR52 pKa = 11.84GSGAVIQDD60 pKa = 3.71SVPLSKK66 pKa = 10.87LSMVIGYY73 pKa = 9.11LLAARR78 pKa = 11.84NIAEE82 pKa = 3.96QLLFIGVSTRR92 pKa = 11.84MSTPLRR98 pKa = 11.84APNGNFLEE106 pKa = 5.13PIVRR110 pKa = 11.84AYY112 pKa = 11.01NFYY115 pKa = 11.14GHH117 pKa = 6.84FEE119 pKa = 4.33HH120 pKa = 7.35EE121 pKa = 4.6SKK123 pKa = 9.93TFVTRR128 pKa = 11.84GLEE131 pKa = 4.09TLFIRR136 pKa = 11.84YY137 pKa = 8.79LFGLRR142 pKa = 11.84QFATTGVSDD151 pKa = 4.61GSAHH155 pKa = 5.0TVRR158 pKa = 11.84VLSNQNHH165 pKa = 5.62NFSDD169 pKa = 3.5IDD171 pKa = 3.86LDD173 pKa = 4.46FYY175 pKa = 11.55YY176 pKa = 10.64LSEE179 pKa = 5.01AGEE182 pKa = 3.99IAFGKK187 pKa = 10.1RR188 pKa = 11.84YY189 pKa = 8.78PLKK192 pKa = 10.01EE193 pKa = 3.95YY194 pKa = 10.48LIHH197 pKa = 5.8LVKK200 pKa = 10.64YY201 pKa = 10.42ASDD204 pKa = 3.57FGDD207 pKa = 3.17VDD209 pKa = 3.4EE210 pKa = 6.36RR211 pKa = 11.84IPLLQRR217 pKa = 11.84ILDD220 pKa = 3.76VTTEE224 pKa = 4.11DD225 pKa = 4.42GLNQFLRR232 pKa = 11.84YY233 pKa = 9.57ARR235 pKa = 11.84RR236 pKa = 11.84LPDD239 pKa = 2.97WQTPPRR245 pKa = 11.84EE246 pKa = 4.16FTPDD250 pKa = 3.22PSDD253 pKa = 3.21KK254 pKa = 10.59HH255 pKa = 6.76KK256 pKa = 11.32SAMKK260 pKa = 9.87EE261 pKa = 3.92LFGAAYY267 pKa = 8.26STAGQSIPVSKK278 pKa = 9.42FTEE281 pKa = 4.71SINLAYY287 pKa = 10.45ALLRR291 pKa = 11.84RR292 pKa = 11.84VSLEE296 pKa = 3.48RR297 pKa = 11.84QYY299 pKa = 12.08GMKK302 pKa = 9.41TVQIPRR308 pKa = 11.84YY309 pKa = 8.61EE310 pKa = 4.43GGSPSQMASYY320 pKa = 10.69QDD322 pKa = 4.16DD323 pKa = 4.1VLFSTIPLSLADD335 pKa = 3.38STAAVGFTTSYY346 pKa = 11.25GSLARR351 pKa = 11.84YY352 pKa = 7.09TSAPGFEE359 pKa = 4.16KK360 pKa = 10.91DD361 pKa = 3.32EE362 pKa = 4.27LVRR365 pKa = 11.84EE366 pKa = 4.33LVSQSLVLRR375 pKa = 4.52
Molecular weight: 41.67 kDa
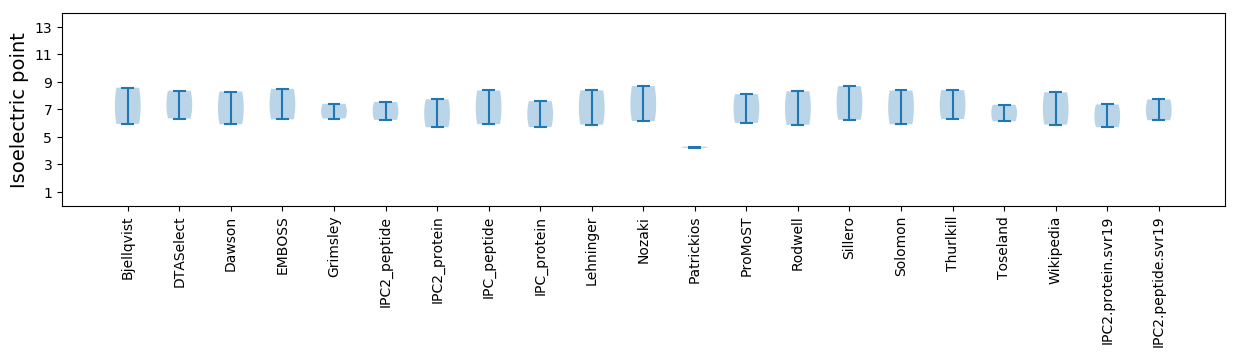
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5M8T3|S5M8T3_9VIRU Uncharacterized protein OS=Ustilaginoidea virens partitivirus 2 OX=1415665 PE=4 SV=1
MM1 pKa = 7.69NSFEE5 pKa = 4.45NFGSFKK11 pKa = 10.93LSADD15 pKa = 3.55EE16 pKa = 4.4LAATAVPPTPWNNVFRR32 pKa = 11.84YY33 pKa = 8.37ITDD36 pKa = 3.46AKK38 pKa = 10.39RR39 pKa = 11.84FPGYY43 pKa = 10.11KK44 pKa = 9.48RR45 pKa = 11.84GILRR49 pKa = 11.84QTQLYY54 pKa = 9.83DD55 pKa = 3.7PYY57 pKa = 11.55VNAALKK63 pKa = 10.55SFSPEE68 pKa = 3.48LHH70 pKa = 7.06DD71 pKa = 5.46SIKK74 pKa = 10.73GYY76 pKa = 8.53TRR78 pKa = 11.84APGDD82 pKa = 3.09EE83 pKa = 3.73WDD85 pKa = 3.64VYY87 pKa = 11.14EE88 pKa = 5.59RR89 pKa = 11.84LTRR92 pKa = 11.84YY93 pKa = 9.63DD94 pKa = 3.87KK95 pKa = 11.54SPLAPVDD102 pKa = 3.38NPRR105 pKa = 11.84FKK107 pKa = 10.92ACYY110 pKa = 9.41DD111 pKa = 3.45AALSDD116 pKa = 3.61VMKK119 pKa = 10.23EE120 pKa = 4.01FKK122 pKa = 10.77LRR124 pKa = 11.84DD125 pKa = 3.48PVVPHH130 pKa = 7.26WILDD134 pKa = 3.42VDD136 pKa = 4.35LVKK139 pKa = 10.22NTSSGFPHH147 pKa = 6.64FTRR150 pKa = 11.84KK151 pKa = 9.94GDD153 pKa = 3.35ILDD156 pKa = 3.89QIRR159 pKa = 11.84QEE161 pKa = 4.32GRR163 pKa = 11.84SHH165 pKa = 6.46FHH167 pKa = 5.97LLKK170 pKa = 10.56RR171 pKa = 11.84LPLWRR176 pKa = 11.84VPLLPCTPATRR187 pKa = 11.84GGLADD192 pKa = 3.06ITEE195 pKa = 4.47PKK197 pKa = 9.07TRR199 pKa = 11.84LVWMYY204 pKa = 8.82PAAMTAVEE212 pKa = 4.79AVFAQPLIDD221 pKa = 4.11GLFSEE226 pKa = 4.64KK227 pKa = 10.41SEE229 pKa = 3.94YY230 pKa = 10.82LITGVDD236 pKa = 3.35TKK238 pKa = 11.16HH239 pKa = 6.82RR240 pKa = 11.84IQRR243 pKa = 11.84YY244 pKa = 8.99LSLLSEE250 pKa = 4.23DD251 pKa = 3.44TGRR254 pKa = 11.84LGVGLDD260 pKa = 3.73FKK262 pKa = 11.63SFDD265 pKa = 3.58TLRR268 pKa = 11.84CNWLIRR274 pKa = 11.84DD275 pKa = 3.82AFDD278 pKa = 3.26VLKK281 pKa = 10.97QNVYY285 pKa = 10.23FSGYY289 pKa = 10.28YY290 pKa = 10.28DD291 pKa = 3.93DD292 pKa = 5.31TNGLQTFGPGKK303 pKa = 8.11TEE305 pKa = 3.93RR306 pKa = 11.84LEE308 pKa = 4.3HH309 pKa = 5.8AWSNIVEE316 pKa = 4.36YY317 pKa = 10.07FIHH320 pKa = 6.56TPILLPNGRR329 pKa = 11.84CVNKK333 pKa = 9.15HH334 pKa = 5.06TGVPSGSHH342 pKa = 4.96FTNLIDD348 pKa = 5.71SIICRR353 pKa = 11.84ILIKK357 pKa = 10.0TFSLYY362 pKa = 10.67CSIPISNLRR371 pKa = 11.84TNGDD375 pKa = 3.28DD376 pKa = 3.77SAFHH380 pKa = 6.83VYY382 pKa = 10.22EE383 pKa = 5.59DD384 pKa = 3.92YY385 pKa = 11.42ASDD388 pKa = 3.95IILRR392 pKa = 11.84AAGFFKK398 pKa = 10.6EE399 pKa = 4.32FFGMTINTDD408 pKa = 3.36KK409 pKa = 11.28SCVAGSPSEE418 pKa = 3.93MHH420 pKa = 6.3VSGTRR425 pKa = 11.84WTGLRR430 pKa = 11.84PTRR433 pKa = 11.84STQEE437 pKa = 3.45WMMLAAYY444 pKa = 9.87GEE446 pKa = 4.64TYY448 pKa = 10.91SRR450 pKa = 11.84IPFDD454 pKa = 3.25SFQRR458 pKa = 11.84LLGLGLSGGFGDD470 pKa = 3.94STFTRR475 pKa = 11.84FFDD478 pKa = 3.97YY479 pKa = 10.6FQTGYY484 pKa = 10.96DD485 pKa = 3.78CRR487 pKa = 11.84HH488 pKa = 6.07GPNLLNWKK496 pKa = 9.6KK497 pKa = 10.64LRR499 pKa = 11.84FLQQIFSIEE508 pKa = 4.12EE509 pKa = 3.82LPLVYY514 pKa = 10.25KK515 pKa = 10.72QGAKK519 pKa = 6.04TTLRR523 pKa = 11.84LRR525 pKa = 11.84LLVTT529 pKa = 4.34
MM1 pKa = 7.69NSFEE5 pKa = 4.45NFGSFKK11 pKa = 10.93LSADD15 pKa = 3.55EE16 pKa = 4.4LAATAVPPTPWNNVFRR32 pKa = 11.84YY33 pKa = 8.37ITDD36 pKa = 3.46AKK38 pKa = 10.39RR39 pKa = 11.84FPGYY43 pKa = 10.11KK44 pKa = 9.48RR45 pKa = 11.84GILRR49 pKa = 11.84QTQLYY54 pKa = 9.83DD55 pKa = 3.7PYY57 pKa = 11.55VNAALKK63 pKa = 10.55SFSPEE68 pKa = 3.48LHH70 pKa = 7.06DD71 pKa = 5.46SIKK74 pKa = 10.73GYY76 pKa = 8.53TRR78 pKa = 11.84APGDD82 pKa = 3.09EE83 pKa = 3.73WDD85 pKa = 3.64VYY87 pKa = 11.14EE88 pKa = 5.59RR89 pKa = 11.84LTRR92 pKa = 11.84YY93 pKa = 9.63DD94 pKa = 3.87KK95 pKa = 11.54SPLAPVDD102 pKa = 3.38NPRR105 pKa = 11.84FKK107 pKa = 10.92ACYY110 pKa = 9.41DD111 pKa = 3.45AALSDD116 pKa = 3.61VMKK119 pKa = 10.23EE120 pKa = 4.01FKK122 pKa = 10.77LRR124 pKa = 11.84DD125 pKa = 3.48PVVPHH130 pKa = 7.26WILDD134 pKa = 3.42VDD136 pKa = 4.35LVKK139 pKa = 10.22NTSSGFPHH147 pKa = 6.64FTRR150 pKa = 11.84KK151 pKa = 9.94GDD153 pKa = 3.35ILDD156 pKa = 3.89QIRR159 pKa = 11.84QEE161 pKa = 4.32GRR163 pKa = 11.84SHH165 pKa = 6.46FHH167 pKa = 5.97LLKK170 pKa = 10.56RR171 pKa = 11.84LPLWRR176 pKa = 11.84VPLLPCTPATRR187 pKa = 11.84GGLADD192 pKa = 3.06ITEE195 pKa = 4.47PKK197 pKa = 9.07TRR199 pKa = 11.84LVWMYY204 pKa = 8.82PAAMTAVEE212 pKa = 4.79AVFAQPLIDD221 pKa = 4.11GLFSEE226 pKa = 4.64KK227 pKa = 10.41SEE229 pKa = 3.94YY230 pKa = 10.82LITGVDD236 pKa = 3.35TKK238 pKa = 11.16HH239 pKa = 6.82RR240 pKa = 11.84IQRR243 pKa = 11.84YY244 pKa = 8.99LSLLSEE250 pKa = 4.23DD251 pKa = 3.44TGRR254 pKa = 11.84LGVGLDD260 pKa = 3.73FKK262 pKa = 11.63SFDD265 pKa = 3.58TLRR268 pKa = 11.84CNWLIRR274 pKa = 11.84DD275 pKa = 3.82AFDD278 pKa = 3.26VLKK281 pKa = 10.97QNVYY285 pKa = 10.23FSGYY289 pKa = 10.28YY290 pKa = 10.28DD291 pKa = 3.93DD292 pKa = 5.31TNGLQTFGPGKK303 pKa = 8.11TEE305 pKa = 3.93RR306 pKa = 11.84LEE308 pKa = 4.3HH309 pKa = 5.8AWSNIVEE316 pKa = 4.36YY317 pKa = 10.07FIHH320 pKa = 6.56TPILLPNGRR329 pKa = 11.84CVNKK333 pKa = 9.15HH334 pKa = 5.06TGVPSGSHH342 pKa = 4.96FTNLIDD348 pKa = 5.71SIICRR353 pKa = 11.84ILIKK357 pKa = 10.0TFSLYY362 pKa = 10.67CSIPISNLRR371 pKa = 11.84TNGDD375 pKa = 3.28DD376 pKa = 3.77SAFHH380 pKa = 6.83VYY382 pKa = 10.22EE383 pKa = 5.59DD384 pKa = 3.92YY385 pKa = 11.42ASDD388 pKa = 3.95IILRR392 pKa = 11.84AAGFFKK398 pKa = 10.6EE399 pKa = 4.32FFGMTINTDD408 pKa = 3.36KK409 pKa = 11.28SCVAGSPSEE418 pKa = 3.93MHH420 pKa = 6.3VSGTRR425 pKa = 11.84WTGLRR430 pKa = 11.84PTRR433 pKa = 11.84STQEE437 pKa = 3.45WMMLAAYY444 pKa = 9.87GEE446 pKa = 4.64TYY448 pKa = 10.91SRR450 pKa = 11.84IPFDD454 pKa = 3.25SFQRR458 pKa = 11.84LLGLGLSGGFGDD470 pKa = 3.94STFTRR475 pKa = 11.84FFDD478 pKa = 3.97YY479 pKa = 10.6FQTGYY484 pKa = 10.96DD485 pKa = 3.78CRR487 pKa = 11.84HH488 pKa = 6.07GPNLLNWKK496 pKa = 9.6KK497 pKa = 10.64LRR499 pKa = 11.84FLQQIFSIEE508 pKa = 4.12EE509 pKa = 3.82LPLVYY514 pKa = 10.25KK515 pKa = 10.72QGAKK519 pKa = 6.04TTLRR523 pKa = 11.84LRR525 pKa = 11.84LLVTT529 pKa = 4.34
Molecular weight: 60.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
904 |
375 |
529 |
452.0 |
51.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.527 ± 0.918 | 0.885 ± 0.551 |
5.973 ± 0.565 | 4.867 ± 0.623 |
6.305 ± 0.273 | 6.858 ± 0.119 |
2.102 ± 0.313 | 4.867 ± 0.208 |
4.314 ± 0.528 | 10.73 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.549 ± 0.032 | 3.208 ± 0.337 |
5.531 ± 0.123 | 3.429 ± 0.688 |
6.637 ± 0.148 | 8.075 ± 1.116 |
6.858 ± 0.286 | 5.642 ± 0.639 |
1.217 ± 0.592 | 4.425 ± 0.068 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
