
Giant panda circovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
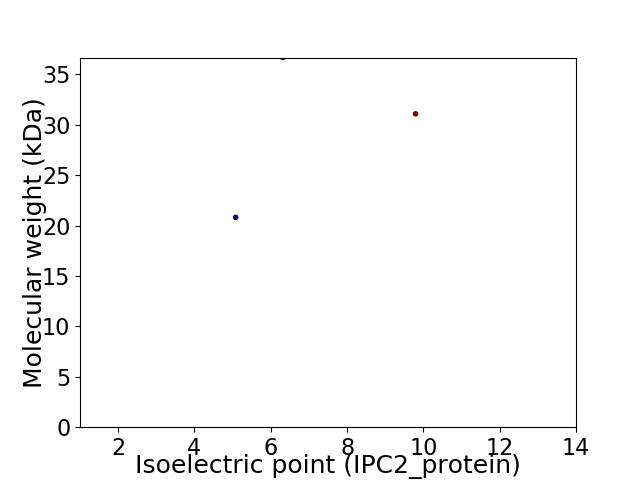
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
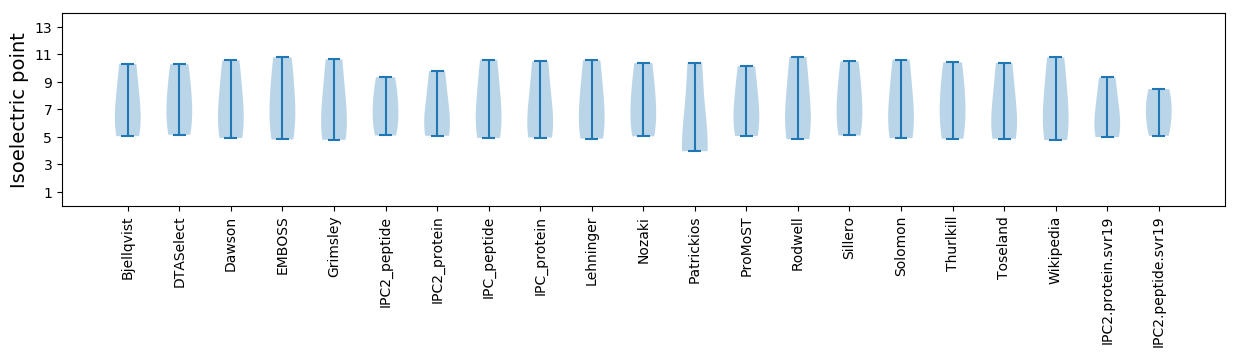
Protein with the lowest isoelectric point:
>tr|A0A220IGS9|A0A220IGS9_9CIRC Capsid protein OS=Giant panda circovirus 2 OX=2016457 PE=4 SV=1
MM1 pKa = 8.15DD2 pKa = 4.36ICLHH6 pKa = 6.28CKK8 pKa = 10.19RR9 pKa = 11.84SPLHH13 pKa = 5.85WRR15 pKa = 11.84WTICSMVEE23 pKa = 3.87NQLPQQDD30 pKa = 3.67SDD32 pKa = 3.21ICRR35 pKa = 11.84AILSSLRR42 pKa = 11.84DD43 pKa = 3.56ADD45 pKa = 3.61STLCEE50 pKa = 4.03RR51 pKa = 11.84CFLEE55 pKa = 3.76HH56 pKa = 6.23TLNVRR61 pKa = 11.84KK62 pKa = 9.83ALTNKK67 pKa = 9.04TMTIAQRR74 pKa = 11.84TEE76 pKa = 3.82TLKK79 pKa = 11.14NLVNSPPTKK88 pKa = 9.38EE89 pKa = 3.63ASVPVFGNLTEE100 pKa = 4.85NGASPWTTSPLIWKK114 pKa = 9.2SLNPFPLCTEE124 pKa = 5.38DD125 pKa = 2.98IAVRR129 pKa = 11.84ADD131 pKa = 3.8LSQTFTALAQQSEE144 pKa = 4.91SEE146 pKa = 4.21SSEE149 pKa = 4.02TGSQSSSSASKK160 pKa = 9.78MSPTIEE166 pKa = 4.04QLNSSSTSLEE176 pKa = 3.81EE177 pKa = 4.31SVSLGSADD185 pKa = 3.39TYY187 pKa = 11.26SANN190 pKa = 3.6
MM1 pKa = 8.15DD2 pKa = 4.36ICLHH6 pKa = 6.28CKK8 pKa = 10.19RR9 pKa = 11.84SPLHH13 pKa = 5.85WRR15 pKa = 11.84WTICSMVEE23 pKa = 3.87NQLPQQDD30 pKa = 3.67SDD32 pKa = 3.21ICRR35 pKa = 11.84AILSSLRR42 pKa = 11.84DD43 pKa = 3.56ADD45 pKa = 3.61STLCEE50 pKa = 4.03RR51 pKa = 11.84CFLEE55 pKa = 3.76HH56 pKa = 6.23TLNVRR61 pKa = 11.84KK62 pKa = 9.83ALTNKK67 pKa = 9.04TMTIAQRR74 pKa = 11.84TEE76 pKa = 3.82TLKK79 pKa = 11.14NLVNSPPTKK88 pKa = 9.38EE89 pKa = 3.63ASVPVFGNLTEE100 pKa = 4.85NGASPWTTSPLIWKK114 pKa = 9.2SLNPFPLCTEE124 pKa = 5.38DD125 pKa = 2.98IAVRR129 pKa = 11.84ADD131 pKa = 3.8LSQTFTALAQQSEE144 pKa = 4.91SEE146 pKa = 4.21SSEE149 pKa = 4.02TGSQSSSSASKK160 pKa = 9.78MSPTIEE166 pKa = 4.04QLNSSSTSLEE176 pKa = 3.81EE177 pKa = 4.31SVSLGSADD185 pKa = 3.39TYY187 pKa = 11.26SANN190 pKa = 3.6
Molecular weight: 20.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A220IGT9|A0A220IGT9_9CIRC Replication-associated protein OS=Giant panda circovirus 2 OX=2016457 PE=4 SV=1
MM1 pKa = 7.5AALVPYY7 pKa = 10.14VGRR10 pKa = 11.84LAMRR14 pKa = 11.84AGLSNPYY21 pKa = 10.2ARR23 pKa = 11.84RR24 pKa = 11.84AGYY27 pKa = 10.8AFAGRR32 pKa = 11.84VAGMAARR39 pKa = 11.84AGMKK43 pKa = 9.57RR44 pKa = 11.84GYY46 pKa = 9.59RR47 pKa = 11.84AVRR50 pKa = 11.84RR51 pKa = 11.84FARR54 pKa = 11.84RR55 pKa = 11.84PRR57 pKa = 11.84AKK59 pKa = 10.02RR60 pKa = 11.84QRR62 pKa = 11.84LNSRR66 pKa = 11.84SHH68 pKa = 6.88VGEE71 pKa = 4.78AIGTTNCKK79 pKa = 8.94TKK81 pKa = 10.38QCSLTDD87 pKa = 3.26ITAKK91 pKa = 10.35NSRR94 pKa = 11.84TLYY97 pKa = 8.95RR98 pKa = 11.84TGVTDD103 pKa = 4.23IPGGSNPDD111 pKa = 2.91EE112 pKa = 4.04RR113 pKa = 11.84LRR115 pKa = 11.84NHH117 pKa = 5.79VNLRR121 pKa = 11.84GFKK124 pKa = 10.13VCLEE128 pKa = 3.81FRR130 pKa = 11.84NNVTTPLYY138 pKa = 11.12VNVAVLAPKK147 pKa = 8.73NTSGNILSEE156 pKa = 3.97TDD158 pKa = 3.71FFRR161 pKa = 11.84DD162 pKa = 3.37SGTTRR167 pKa = 11.84AKK169 pKa = 10.76DD170 pKa = 3.35FSTALTAMEE179 pKa = 4.42FHH181 pKa = 7.1CLPVNTDD188 pKa = 3.48DD189 pKa = 4.92YY190 pKa = 12.07VVLKK194 pKa = 10.28HH195 pKa = 6.2KK196 pKa = 10.51RR197 pKa = 11.84FRR199 pKa = 11.84LGPLPGATLYY209 pKa = 10.88QNSVGNNYY217 pKa = 10.23KK218 pKa = 10.59NVDD221 pKa = 3.12WYY223 pKa = 10.42IKK225 pKa = 10.13LGRR228 pKa = 11.84QVRR231 pKa = 11.84YY232 pKa = 9.44EE233 pKa = 4.14SSSSQATDD241 pKa = 2.89GHH243 pKa = 6.74VSVAYY248 pKa = 9.15WCDD251 pKa = 2.78AWGAAAGTAAASGSMSVSQRR271 pKa = 11.84YY272 pKa = 4.83ITYY275 pKa = 8.96WKK277 pKa = 10.05EE278 pKa = 3.66PKK280 pKa = 9.02TT281 pKa = 3.79
MM1 pKa = 7.5AALVPYY7 pKa = 10.14VGRR10 pKa = 11.84LAMRR14 pKa = 11.84AGLSNPYY21 pKa = 10.2ARR23 pKa = 11.84RR24 pKa = 11.84AGYY27 pKa = 10.8AFAGRR32 pKa = 11.84VAGMAARR39 pKa = 11.84AGMKK43 pKa = 9.57RR44 pKa = 11.84GYY46 pKa = 9.59RR47 pKa = 11.84AVRR50 pKa = 11.84RR51 pKa = 11.84FARR54 pKa = 11.84RR55 pKa = 11.84PRR57 pKa = 11.84AKK59 pKa = 10.02RR60 pKa = 11.84QRR62 pKa = 11.84LNSRR66 pKa = 11.84SHH68 pKa = 6.88VGEE71 pKa = 4.78AIGTTNCKK79 pKa = 8.94TKK81 pKa = 10.38QCSLTDD87 pKa = 3.26ITAKK91 pKa = 10.35NSRR94 pKa = 11.84TLYY97 pKa = 8.95RR98 pKa = 11.84TGVTDD103 pKa = 4.23IPGGSNPDD111 pKa = 2.91EE112 pKa = 4.04RR113 pKa = 11.84LRR115 pKa = 11.84NHH117 pKa = 5.79VNLRR121 pKa = 11.84GFKK124 pKa = 10.13VCLEE128 pKa = 3.81FRR130 pKa = 11.84NNVTTPLYY138 pKa = 11.12VNVAVLAPKK147 pKa = 8.73NTSGNILSEE156 pKa = 3.97TDD158 pKa = 3.71FFRR161 pKa = 11.84DD162 pKa = 3.37SGTTRR167 pKa = 11.84AKK169 pKa = 10.76DD170 pKa = 3.35FSTALTAMEE179 pKa = 4.42FHH181 pKa = 7.1CLPVNTDD188 pKa = 3.48DD189 pKa = 4.92YY190 pKa = 12.07VVLKK194 pKa = 10.28HH195 pKa = 6.2KK196 pKa = 10.51RR197 pKa = 11.84FRR199 pKa = 11.84LGPLPGATLYY209 pKa = 10.88QNSVGNNYY217 pKa = 10.23KK218 pKa = 10.59NVDD221 pKa = 3.12WYY223 pKa = 10.42IKK225 pKa = 10.13LGRR228 pKa = 11.84QVRR231 pKa = 11.84YY232 pKa = 9.44EE233 pKa = 4.14SSSSQATDD241 pKa = 2.89GHH243 pKa = 6.74VSVAYY248 pKa = 9.15WCDD251 pKa = 2.78AWGAAAGTAAASGSMSVSQRR271 pKa = 11.84YY272 pKa = 4.83ITYY275 pKa = 8.96WKK277 pKa = 10.05EE278 pKa = 3.66PKK280 pKa = 9.02TT281 pKa = 3.79
Molecular weight: 31.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
792 |
190 |
321 |
264.0 |
29.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.081 ± 1.555 | 2.904 ± 0.508 |
5.177 ± 0.845 | 5.177 ± 1.22 |
3.409 ± 0.473 | 6.313 ± 1.201 |
2.273 ± 0.41 | 3.157 ± 0.466 |
5.051 ± 0.45 | 9.217 ± 1.126 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.399 ± 0.199 | 4.293 ± 1.092 |
4.672 ± 0.341 | 3.03 ± 0.51 |
7.576 ± 1.29 | 9.343 ± 1.921 |
6.439 ± 1.604 | 6.061 ± 0.748 |
1.641 ± 0.134 | 3.788 ± 0.905 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
