
Novosphingobium kunmingense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
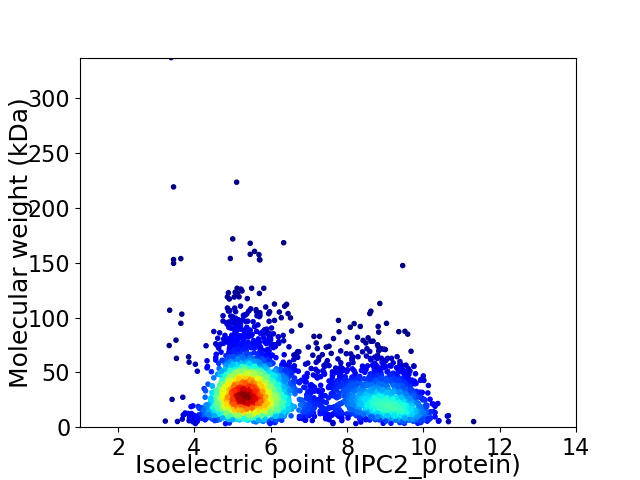
Virtual 2D-PAGE plot for 3342 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
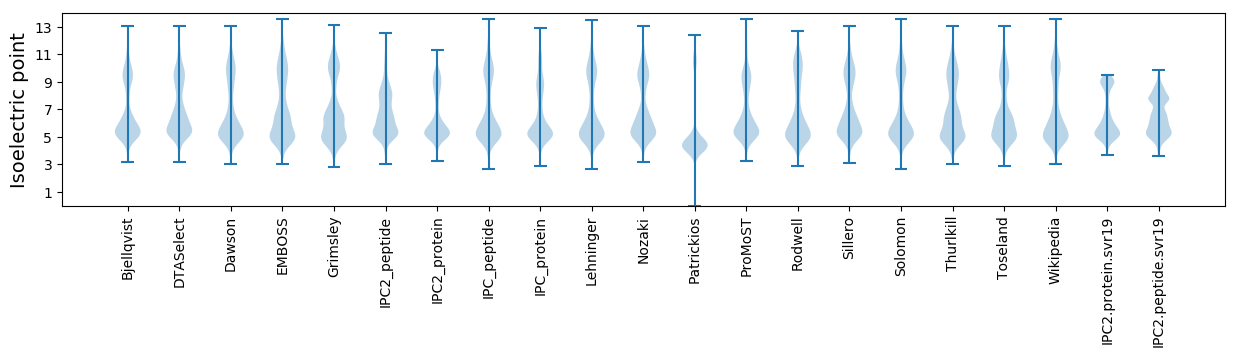
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N0H306|A0A2N0H306_9SPHN NADPH:quinone reductase-like Zn-dependent oxidoreductase OS=Novosphingobium kunmingense OX=1211806 GN=B0I00_3346 PE=4 SV=1
MM1 pKa = 7.16AQFWHH6 pKa = 6.34NFAWWYY12 pKa = 9.77SLPITITDD20 pKa = 3.42TSGNDD25 pKa = 3.76AISGTDD31 pKa = 3.53GADD34 pKa = 3.47HH35 pKa = 7.14ISGNSGDD42 pKa = 4.99DD43 pKa = 3.75EE44 pKa = 4.36IAGLGGDD51 pKa = 4.82DD52 pKa = 3.71YY53 pKa = 11.97LEE55 pKa = 4.7GGNEE59 pKa = 3.6IDD61 pKa = 3.64TLYY64 pKa = 11.27GGAGNDD70 pKa = 3.4MLDD73 pKa = 3.63PGRR76 pKa = 11.84DD77 pKa = 3.0QDD79 pKa = 4.02TVYY82 pKa = 10.96GGPDD86 pKa = 3.24DD87 pKa = 5.7DD88 pKa = 5.18YY89 pKa = 11.31YY90 pKa = 11.48LIYY93 pKa = 10.3GAEE96 pKa = 4.08AAAQDD101 pKa = 4.17TLIEE105 pKa = 4.2AANEE109 pKa = 3.82GFDD112 pKa = 4.37WIRR115 pKa = 11.84SSQDD119 pKa = 2.91GYY121 pKa = 11.48GLPFNFEE128 pKa = 4.12GLFLHH133 pKa = 7.1IDD135 pKa = 3.59GGGGLAYY142 pKa = 10.36DD143 pKa = 4.21GSGNEE148 pKa = 4.1LDD150 pKa = 3.96NVIVGQGLANHH161 pKa = 6.8IFGLGGNDD169 pKa = 3.51LLLGEE174 pKa = 5.4GGNDD178 pKa = 3.2WLDD181 pKa = 3.76GGSGSDD187 pKa = 3.12WLEE190 pKa = 4.07GGLGSDD196 pKa = 3.49WFVGSTGNDD205 pKa = 2.83VMLGDD210 pKa = 4.64GGSDD214 pKa = 3.28TFDD217 pKa = 3.13LRR219 pKa = 11.84AVTFVAGQSVLIDD232 pKa = 3.78GAGSTGDD239 pKa = 3.74PASPDD244 pKa = 2.97VDD246 pKa = 3.64TLVLTGRR253 pKa = 11.84ATDD256 pKa = 3.36WTFTTTFEE264 pKa = 5.92DD265 pKa = 4.8GVQPTRR271 pKa = 11.84TVLTNNLTGTLLYY284 pKa = 9.01TQHH287 pKa = 7.64LEE289 pKa = 4.13ALSFTAPVNNLVNLYY304 pKa = 9.9QGNLVLEE311 pKa = 5.22LIRR314 pKa = 11.84LAGEE318 pKa = 4.13SYY320 pKa = 11.01GALEE324 pKa = 4.36TLDD327 pKa = 4.72HH328 pKa = 6.43LAEE331 pKa = 4.71PLASTGTFAAWAAAGVAAEE350 pKa = 4.23ATQLRR355 pKa = 11.84HH356 pKa = 4.12WHH358 pKa = 6.31ALSALEE364 pKa = 4.3LGIPAAYY371 pKa = 7.7THH373 pKa = 6.81PQTGLQYY380 pKa = 11.25SLTNGHH386 pKa = 5.76YY387 pKa = 9.32QAIYY391 pKa = 8.31PSPVGIVDD399 pKa = 3.57TSEE402 pKa = 4.18ANALVLIGVVNGVTTLALAFRR423 pKa = 11.84GTDD426 pKa = 3.25QFADD430 pKa = 3.78FADD433 pKa = 4.25YY434 pKa = 11.03PDD436 pKa = 3.82FTLDD440 pKa = 3.4HH441 pKa = 5.89YY442 pKa = 11.97AKK444 pKa = 10.46FKK446 pKa = 10.61PLIDD450 pKa = 5.58AIDD453 pKa = 4.37AYY455 pKa = 11.32VADD458 pKa = 4.71PANGIQQVLVSGHH471 pKa = 5.48SLGAGMVQEE480 pKa = 4.88FMSDD484 pKa = 3.07HH485 pKa = 6.67AGSLYY490 pKa = 10.14QAVTIGSPGSDD501 pKa = 3.39NNDD504 pKa = 3.81GIEE507 pKa = 4.33DD508 pKa = 3.84DD509 pKa = 3.76PRR511 pKa = 11.84IVNFAHH517 pKa = 6.43TSDD520 pKa = 4.1AVAMIAPALSQAAPILSAWLLLQGQVTLAAIIGKK554 pKa = 8.46IQPKK558 pKa = 10.34DD559 pKa = 3.47RR560 pKa = 11.84DD561 pKa = 3.63GSDD564 pKa = 3.32VYY566 pKa = 11.54VNSDD570 pKa = 2.55IATFPGITEE579 pKa = 4.33HH580 pKa = 7.2AWTNYY585 pKa = 8.61LASVTTILEE594 pKa = 4.19RR595 pKa = 11.84AVTGPFSLHH604 pKa = 6.3PFAAALRR611 pKa = 11.84AEE613 pKa = 4.48LVYY616 pKa = 11.12SGDD619 pKa = 3.9DD620 pKa = 3.36VQIAIGQPGSSKK632 pKa = 8.56ITSWTGDD639 pKa = 3.3NFVLGSSAADD649 pKa = 3.51SIEE652 pKa = 3.96WSGVTPLGEE661 pKa = 4.07YY662 pKa = 10.06RR663 pKa = 11.84VIDD666 pKa = 3.9AGLGSDD672 pKa = 3.96EE673 pKa = 5.87LILPGLKK680 pKa = 10.43ASWGWDD686 pKa = 3.21NLGGHH691 pKa = 6.11YY692 pKa = 10.65ALSFLGLDD700 pKa = 3.93VGEE703 pKa = 4.28LWNVEE708 pKa = 3.93KK709 pKa = 10.95LVFTFFSDD717 pKa = 4.94EE718 pKa = 4.05EE719 pKa = 4.13LQQAKK724 pKa = 8.18TAFAAAEE731 pKa = 4.15PAPVIPVVYY740 pKa = 10.48LNGQAASVQQAVPGASVLNVDD761 pKa = 3.81PDD763 pKa = 3.27VDD765 pKa = 4.03YY766 pKa = 11.94VEE768 pKa = 5.02TGNSTLMVNGSADD781 pKa = 3.26NDD783 pKa = 3.8IIMLGGGNQTIDD795 pKa = 3.12GGAGNDD801 pKa = 3.29VVNAVNAASGGVFAVTGGGGDD822 pKa = 3.88DD823 pKa = 4.05LVIGSGGARR832 pKa = 11.84MKK834 pKa = 10.84AVFSGNAADD843 pKa = 3.93YY844 pKa = 10.72GADD847 pKa = 3.37EE848 pKa = 5.24RR849 pKa = 11.84GDD851 pKa = 3.5GGIILTDD858 pKa = 4.24LRR860 pKa = 11.84PGSPDD865 pKa = 3.6GEE867 pKa = 4.24DD868 pKa = 2.97TLLQVVTLRR877 pKa = 11.84FADD880 pKa = 3.75GDD882 pKa = 3.47ITAAQFLAEE891 pKa = 4.39RR892 pKa = 11.84VPVLLQGTDD901 pKa = 3.9DD902 pKa = 5.18DD903 pKa = 5.1DD904 pKa = 5.83ALIPARR910 pKa = 11.84DD911 pKa = 3.73GTVIAIGLAGDD922 pKa = 4.27DD923 pKa = 4.06SLEE926 pKa = 4.28GGARR930 pKa = 11.84ADD932 pKa = 4.21LLDD935 pKa = 4.33GGTGADD941 pKa = 3.62HH942 pKa = 7.43LYY944 pKa = 10.92GHH946 pKa = 7.38AGDD949 pKa = 5.92DD950 pKa = 3.71MYY952 pKa = 11.62LVNQLADD959 pKa = 3.56TVFEE963 pKa = 4.58GDD965 pKa = 3.77GEE967 pKa = 4.55GTDD970 pKa = 3.47TVTASGNFYY979 pKa = 10.77LYY981 pKa = 11.23ANIEE985 pKa = 4.22NLVQAAGAA993 pKa = 3.7
MM1 pKa = 7.16AQFWHH6 pKa = 6.34NFAWWYY12 pKa = 9.77SLPITITDD20 pKa = 3.42TSGNDD25 pKa = 3.76AISGTDD31 pKa = 3.53GADD34 pKa = 3.47HH35 pKa = 7.14ISGNSGDD42 pKa = 4.99DD43 pKa = 3.75EE44 pKa = 4.36IAGLGGDD51 pKa = 4.82DD52 pKa = 3.71YY53 pKa = 11.97LEE55 pKa = 4.7GGNEE59 pKa = 3.6IDD61 pKa = 3.64TLYY64 pKa = 11.27GGAGNDD70 pKa = 3.4MLDD73 pKa = 3.63PGRR76 pKa = 11.84DD77 pKa = 3.0QDD79 pKa = 4.02TVYY82 pKa = 10.96GGPDD86 pKa = 3.24DD87 pKa = 5.7DD88 pKa = 5.18YY89 pKa = 11.31YY90 pKa = 11.48LIYY93 pKa = 10.3GAEE96 pKa = 4.08AAAQDD101 pKa = 4.17TLIEE105 pKa = 4.2AANEE109 pKa = 3.82GFDD112 pKa = 4.37WIRR115 pKa = 11.84SSQDD119 pKa = 2.91GYY121 pKa = 11.48GLPFNFEE128 pKa = 4.12GLFLHH133 pKa = 7.1IDD135 pKa = 3.59GGGGLAYY142 pKa = 10.36DD143 pKa = 4.21GSGNEE148 pKa = 4.1LDD150 pKa = 3.96NVIVGQGLANHH161 pKa = 6.8IFGLGGNDD169 pKa = 3.51LLLGEE174 pKa = 5.4GGNDD178 pKa = 3.2WLDD181 pKa = 3.76GGSGSDD187 pKa = 3.12WLEE190 pKa = 4.07GGLGSDD196 pKa = 3.49WFVGSTGNDD205 pKa = 2.83VMLGDD210 pKa = 4.64GGSDD214 pKa = 3.28TFDD217 pKa = 3.13LRR219 pKa = 11.84AVTFVAGQSVLIDD232 pKa = 3.78GAGSTGDD239 pKa = 3.74PASPDD244 pKa = 2.97VDD246 pKa = 3.64TLVLTGRR253 pKa = 11.84ATDD256 pKa = 3.36WTFTTTFEE264 pKa = 5.92DD265 pKa = 4.8GVQPTRR271 pKa = 11.84TVLTNNLTGTLLYY284 pKa = 9.01TQHH287 pKa = 7.64LEE289 pKa = 4.13ALSFTAPVNNLVNLYY304 pKa = 9.9QGNLVLEE311 pKa = 5.22LIRR314 pKa = 11.84LAGEE318 pKa = 4.13SYY320 pKa = 11.01GALEE324 pKa = 4.36TLDD327 pKa = 4.72HH328 pKa = 6.43LAEE331 pKa = 4.71PLASTGTFAAWAAAGVAAEE350 pKa = 4.23ATQLRR355 pKa = 11.84HH356 pKa = 4.12WHH358 pKa = 6.31ALSALEE364 pKa = 4.3LGIPAAYY371 pKa = 7.7THH373 pKa = 6.81PQTGLQYY380 pKa = 11.25SLTNGHH386 pKa = 5.76YY387 pKa = 9.32QAIYY391 pKa = 8.31PSPVGIVDD399 pKa = 3.57TSEE402 pKa = 4.18ANALVLIGVVNGVTTLALAFRR423 pKa = 11.84GTDD426 pKa = 3.25QFADD430 pKa = 3.78FADD433 pKa = 4.25YY434 pKa = 11.03PDD436 pKa = 3.82FTLDD440 pKa = 3.4HH441 pKa = 5.89YY442 pKa = 11.97AKK444 pKa = 10.46FKK446 pKa = 10.61PLIDD450 pKa = 5.58AIDD453 pKa = 4.37AYY455 pKa = 11.32VADD458 pKa = 4.71PANGIQQVLVSGHH471 pKa = 5.48SLGAGMVQEE480 pKa = 4.88FMSDD484 pKa = 3.07HH485 pKa = 6.67AGSLYY490 pKa = 10.14QAVTIGSPGSDD501 pKa = 3.39NNDD504 pKa = 3.81GIEE507 pKa = 4.33DD508 pKa = 3.84DD509 pKa = 3.76PRR511 pKa = 11.84IVNFAHH517 pKa = 6.43TSDD520 pKa = 4.1AVAMIAPALSQAAPILSAWLLLQGQVTLAAIIGKK554 pKa = 8.46IQPKK558 pKa = 10.34DD559 pKa = 3.47RR560 pKa = 11.84DD561 pKa = 3.63GSDD564 pKa = 3.32VYY566 pKa = 11.54VNSDD570 pKa = 2.55IATFPGITEE579 pKa = 4.33HH580 pKa = 7.2AWTNYY585 pKa = 8.61LASVTTILEE594 pKa = 4.19RR595 pKa = 11.84AVTGPFSLHH604 pKa = 6.3PFAAALRR611 pKa = 11.84AEE613 pKa = 4.48LVYY616 pKa = 11.12SGDD619 pKa = 3.9DD620 pKa = 3.36VQIAIGQPGSSKK632 pKa = 8.56ITSWTGDD639 pKa = 3.3NFVLGSSAADD649 pKa = 3.51SIEE652 pKa = 3.96WSGVTPLGEE661 pKa = 4.07YY662 pKa = 10.06RR663 pKa = 11.84VIDD666 pKa = 3.9AGLGSDD672 pKa = 3.96EE673 pKa = 5.87LILPGLKK680 pKa = 10.43ASWGWDD686 pKa = 3.21NLGGHH691 pKa = 6.11YY692 pKa = 10.65ALSFLGLDD700 pKa = 3.93VGEE703 pKa = 4.28LWNVEE708 pKa = 3.93KK709 pKa = 10.95LVFTFFSDD717 pKa = 4.94EE718 pKa = 4.05EE719 pKa = 4.13LQQAKK724 pKa = 8.18TAFAAAEE731 pKa = 4.15PAPVIPVVYY740 pKa = 10.48LNGQAASVQQAVPGASVLNVDD761 pKa = 3.81PDD763 pKa = 3.27VDD765 pKa = 4.03YY766 pKa = 11.94VEE768 pKa = 5.02TGNSTLMVNGSADD781 pKa = 3.26NDD783 pKa = 3.8IIMLGGGNQTIDD795 pKa = 3.12GGAGNDD801 pKa = 3.29VVNAVNAASGGVFAVTGGGGDD822 pKa = 3.88DD823 pKa = 4.05LVIGSGGARR832 pKa = 11.84MKK834 pKa = 10.84AVFSGNAADD843 pKa = 3.93YY844 pKa = 10.72GADD847 pKa = 3.37EE848 pKa = 5.24RR849 pKa = 11.84GDD851 pKa = 3.5GGIILTDD858 pKa = 4.24LRR860 pKa = 11.84PGSPDD865 pKa = 3.6GEE867 pKa = 4.24DD868 pKa = 2.97TLLQVVTLRR877 pKa = 11.84FADD880 pKa = 3.75GDD882 pKa = 3.47ITAAQFLAEE891 pKa = 4.39RR892 pKa = 11.84VPVLLQGTDD901 pKa = 3.9DD902 pKa = 5.18DD903 pKa = 5.1DD904 pKa = 5.83ALIPARR910 pKa = 11.84DD911 pKa = 3.73GTVIAIGLAGDD922 pKa = 4.27DD923 pKa = 4.06SLEE926 pKa = 4.28GGARR930 pKa = 11.84ADD932 pKa = 4.21LLDD935 pKa = 4.33GGTGADD941 pKa = 3.62HH942 pKa = 7.43LYY944 pKa = 10.92GHH946 pKa = 7.38AGDD949 pKa = 5.92DD950 pKa = 3.71MYY952 pKa = 11.62LVNQLADD959 pKa = 3.56TVFEE963 pKa = 4.58GDD965 pKa = 3.77GEE967 pKa = 4.55GTDD970 pKa = 3.47TVTASGNFYY979 pKa = 10.77LYY981 pKa = 11.23ANIEE985 pKa = 4.22NLVQAAGAA993 pKa = 3.7
Molecular weight: 103.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N0H6X2|A0A2N0H6X2_9SPHN Na+/H+-dicarboxylate symporter OS=Novosphingobium kunmingense OX=1211806 GN=B0I00_2286 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 8.17VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 8.17VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1082236 |
29 |
3423 |
323.8 |
34.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.603 ± 0.07 | 0.844 ± 0.013 |
6.071 ± 0.042 | 5.337 ± 0.039 |
3.543 ± 0.025 | 9.034 ± 0.04 |
1.945 ± 0.021 | 4.724 ± 0.027 |
2.969 ± 0.03 | 10.02 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.366 ± 0.022 | 2.53 ± 0.038 |
5.316 ± 0.03 | 3.114 ± 0.021 |
7.237 ± 0.051 | 5.181 ± 0.03 |
5.245 ± 0.048 | 7.266 ± 0.033 |
1.482 ± 0.018 | 2.174 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
