
Paracoccus siganidrum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
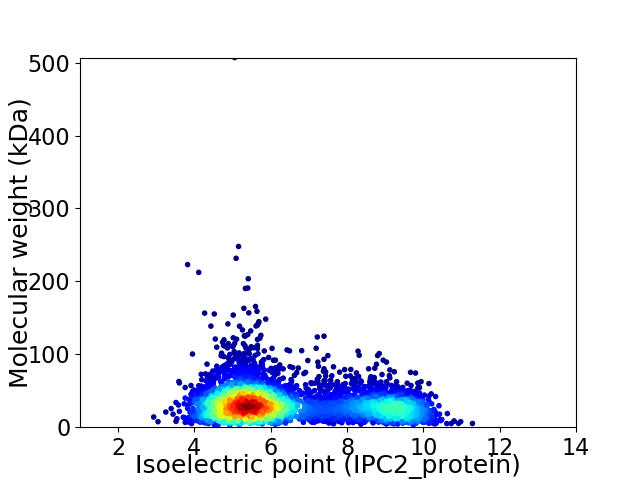
Virtual 2D-PAGE plot for 4665 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A418ZT36|A0A418ZT36_9RHOB Methylamine utilization protein MauF (Fragment) OS=Paracoccus siganidrum OX=1276757 GN=D3P05_23105 PE=4 SV=1
MM1 pKa = 7.51GAGSDD6 pKa = 3.89PVCAGNARR14 pKa = 11.84KK15 pKa = 9.7RR16 pKa = 11.84KK17 pKa = 7.98EE18 pKa = 3.88LEE20 pKa = 3.86MPIGTNFNDD29 pKa = 3.91TLIGDD34 pKa = 3.76NTAEE38 pKa = 4.3LFFGLGGNDD47 pKa = 4.43LILGNGGNDD56 pKa = 3.61TIYY59 pKa = 11.06GGTGNDD65 pKa = 3.62TVDD68 pKa = 3.45GGSGNDD74 pKa = 4.11LIYY77 pKa = 11.27GEE79 pKa = 5.22AGNDD83 pKa = 3.57SLVGGGGTDD92 pKa = 3.6TIYY95 pKa = 11.02GGEE98 pKa = 4.2GNDD101 pKa = 4.08TIRR104 pKa = 11.84SSGAGFYY111 pKa = 10.9DD112 pKa = 3.87GGNGNDD118 pKa = 3.43YY119 pKa = 10.88VYY121 pKa = 11.06AGLGVAEE128 pKa = 4.45TLRR131 pKa = 11.84GGAGVDD137 pKa = 3.35WLNTTNWNSAYY148 pKa = 10.44VINLATGLTNFAGEE162 pKa = 4.4SFTQFEE168 pKa = 4.46NLVTGNGNDD177 pKa = 3.87SLTGTADD184 pKa = 3.05ANTIYY189 pKa = 10.72SGGGDD194 pKa = 3.17DD195 pKa = 3.96TVRR198 pKa = 11.84GEE200 pKa = 4.41AGNDD204 pKa = 3.5YY205 pKa = 10.73ISAGGGNDD213 pKa = 3.65LLYY216 pKa = 10.93GGSGNDD222 pKa = 3.33TVLGGDD228 pKa = 3.82GNDD231 pKa = 3.21RR232 pKa = 11.84LYY234 pKa = 11.54GEE236 pKa = 5.11AGNDD240 pKa = 3.53YY241 pKa = 10.99LYY243 pKa = 11.49GDD245 pKa = 5.01AGNDD249 pKa = 3.88TIDD252 pKa = 3.72GGSGNDD258 pKa = 3.61YY259 pKa = 11.09LSGGSGNDD267 pKa = 3.2SLTGGGGTDD276 pKa = 3.65TIYY279 pKa = 11.02GGEE282 pKa = 4.2GNDD285 pKa = 4.08TIRR288 pKa = 11.84SSGAGVYY295 pKa = 10.18DD296 pKa = 3.93GQAGDD301 pKa = 3.64DD302 pKa = 3.6HH303 pKa = 8.75VYY305 pKa = 11.08AGLGVAEE312 pKa = 4.45TLRR315 pKa = 11.84GGAGVDD321 pKa = 3.07WLDD324 pKa = 3.26TTTYY328 pKa = 9.93GGNYY332 pKa = 9.35QVDD335 pKa = 4.23LASGLTNYY343 pKa = 10.31AGEE346 pKa = 4.32SFVQFDD352 pKa = 4.5HH353 pKa = 6.45LQSGQGHH360 pKa = 6.25DD361 pKa = 3.95TLFGTAGDD369 pKa = 3.75NQIRR373 pKa = 11.84GGGGADD379 pKa = 3.86RR380 pKa = 11.84IHH382 pKa = 6.52GRR384 pKa = 11.84AGHH387 pKa = 6.97DD388 pKa = 3.9LLNGQAGHH396 pKa = 6.92DD397 pKa = 3.78TLNGNNGNDD406 pKa = 3.71TLLGQAGNDD415 pKa = 3.69VLNGGAGQDD424 pKa = 3.75VLRR427 pKa = 11.84GGAGSDD433 pKa = 3.18ILSGQQGHH441 pKa = 6.41DD442 pKa = 3.4QLFGEE447 pKa = 5.16AGPDD451 pKa = 3.43QLLGGAGNDD460 pKa = 3.89TLNGGAGHH468 pKa = 6.92DD469 pKa = 4.11TLNGGAGNDD478 pKa = 4.03LLIGGAGNDD487 pKa = 3.82LLIGGGGEE495 pKa = 3.86DD496 pKa = 3.16TFRR499 pKa = 11.84FVSISDD505 pKa = 3.75SPSGGNHH512 pKa = 5.39DD513 pKa = 4.85TISGFQGAGNRR524 pKa = 11.84PAAAIEE530 pKa = 4.0DD531 pKa = 4.91LIDD534 pKa = 5.08LSQIDD539 pKa = 4.37ANTLVGGNQAFTFNGTTPGGVGTIWLQNVGNATWLHH575 pKa = 6.15LNVDD579 pKa = 3.42NDD581 pKa = 3.96AAPEE585 pKa = 3.91MTIRR589 pKa = 11.84ILDD592 pKa = 3.95GATTAGDD599 pKa = 3.44YY600 pKa = 8.55WAGDD604 pKa = 4.14FILL607 pKa = 5.43
MM1 pKa = 7.51GAGSDD6 pKa = 3.89PVCAGNARR14 pKa = 11.84KK15 pKa = 9.7RR16 pKa = 11.84KK17 pKa = 7.98EE18 pKa = 3.88LEE20 pKa = 3.86MPIGTNFNDD29 pKa = 3.91TLIGDD34 pKa = 3.76NTAEE38 pKa = 4.3LFFGLGGNDD47 pKa = 4.43LILGNGGNDD56 pKa = 3.61TIYY59 pKa = 11.06GGTGNDD65 pKa = 3.62TVDD68 pKa = 3.45GGSGNDD74 pKa = 4.11LIYY77 pKa = 11.27GEE79 pKa = 5.22AGNDD83 pKa = 3.57SLVGGGGTDD92 pKa = 3.6TIYY95 pKa = 11.02GGEE98 pKa = 4.2GNDD101 pKa = 4.08TIRR104 pKa = 11.84SSGAGFYY111 pKa = 10.9DD112 pKa = 3.87GGNGNDD118 pKa = 3.43YY119 pKa = 10.88VYY121 pKa = 11.06AGLGVAEE128 pKa = 4.45TLRR131 pKa = 11.84GGAGVDD137 pKa = 3.35WLNTTNWNSAYY148 pKa = 10.44VINLATGLTNFAGEE162 pKa = 4.4SFTQFEE168 pKa = 4.46NLVTGNGNDD177 pKa = 3.87SLTGTADD184 pKa = 3.05ANTIYY189 pKa = 10.72SGGGDD194 pKa = 3.17DD195 pKa = 3.96TVRR198 pKa = 11.84GEE200 pKa = 4.41AGNDD204 pKa = 3.5YY205 pKa = 10.73ISAGGGNDD213 pKa = 3.65LLYY216 pKa = 10.93GGSGNDD222 pKa = 3.33TVLGGDD228 pKa = 3.82GNDD231 pKa = 3.21RR232 pKa = 11.84LYY234 pKa = 11.54GEE236 pKa = 5.11AGNDD240 pKa = 3.53YY241 pKa = 10.99LYY243 pKa = 11.49GDD245 pKa = 5.01AGNDD249 pKa = 3.88TIDD252 pKa = 3.72GGSGNDD258 pKa = 3.61YY259 pKa = 11.09LSGGSGNDD267 pKa = 3.2SLTGGGGTDD276 pKa = 3.65TIYY279 pKa = 11.02GGEE282 pKa = 4.2GNDD285 pKa = 4.08TIRR288 pKa = 11.84SSGAGVYY295 pKa = 10.18DD296 pKa = 3.93GQAGDD301 pKa = 3.64DD302 pKa = 3.6HH303 pKa = 8.75VYY305 pKa = 11.08AGLGVAEE312 pKa = 4.45TLRR315 pKa = 11.84GGAGVDD321 pKa = 3.07WLDD324 pKa = 3.26TTTYY328 pKa = 9.93GGNYY332 pKa = 9.35QVDD335 pKa = 4.23LASGLTNYY343 pKa = 10.31AGEE346 pKa = 4.32SFVQFDD352 pKa = 4.5HH353 pKa = 6.45LQSGQGHH360 pKa = 6.25DD361 pKa = 3.95TLFGTAGDD369 pKa = 3.75NQIRR373 pKa = 11.84GGGGADD379 pKa = 3.86RR380 pKa = 11.84IHH382 pKa = 6.52GRR384 pKa = 11.84AGHH387 pKa = 6.97DD388 pKa = 3.9LLNGQAGHH396 pKa = 6.92DD397 pKa = 3.78TLNGNNGNDD406 pKa = 3.71TLLGQAGNDD415 pKa = 3.69VLNGGAGQDD424 pKa = 3.75VLRR427 pKa = 11.84GGAGSDD433 pKa = 3.18ILSGQQGHH441 pKa = 6.41DD442 pKa = 3.4QLFGEE447 pKa = 5.16AGPDD451 pKa = 3.43QLLGGAGNDD460 pKa = 3.89TLNGGAGHH468 pKa = 6.92DD469 pKa = 4.11TLNGGAGNDD478 pKa = 4.03LLIGGAGNDD487 pKa = 3.82LLIGGGGEE495 pKa = 3.86DD496 pKa = 3.16TFRR499 pKa = 11.84FVSISDD505 pKa = 3.75SPSGGNHH512 pKa = 5.39DD513 pKa = 4.85TISGFQGAGNRR524 pKa = 11.84PAAAIEE530 pKa = 4.0DD531 pKa = 4.91LIDD534 pKa = 5.08LSQIDD539 pKa = 4.37ANTLVGGNQAFTFNGTTPGGVGTIWLQNVGNATWLHH575 pKa = 6.15LNVDD579 pKa = 3.42NDD581 pKa = 3.96AAPEE585 pKa = 3.91MTIRR589 pKa = 11.84ILDD592 pKa = 3.95GATTAGDD599 pKa = 3.44YY600 pKa = 8.55WAGDD604 pKa = 4.14FILL607 pKa = 5.43
Molecular weight: 60.68 kDa
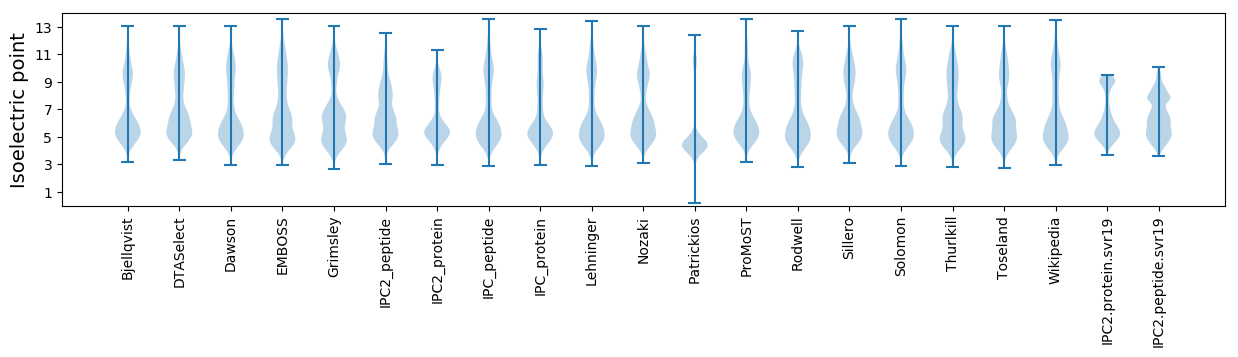
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A419A9J6|A0A419A9J6_9RHOB DUF3445 domain-containing protein OS=Paracoccus siganidrum OX=1276757 GN=D3P05_05035 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12GGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.69GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12GGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.69GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1441716 |
28 |
4715 |
309.0 |
33.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.43 ± 0.057 | 0.85 ± 0.012 |
5.89 ± 0.031 | 5.665 ± 0.031 |
3.507 ± 0.022 | 9.15 ± 0.039 |
2.117 ± 0.017 | 5.066 ± 0.023 |
2.276 ± 0.029 | 10.475 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.7 ± 0.017 | 2.25 ± 0.021 |
5.566 ± 0.029 | 3.26 ± 0.02 |
7.818 ± 0.039 | 4.745 ± 0.023 |
4.881 ± 0.026 | 6.916 ± 0.026 |
1.466 ± 0.016 | 1.974 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
