
Azoarcus sp. (strain BH72)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Rhodocyclales; Zoogloeaceae; Azoarcus
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
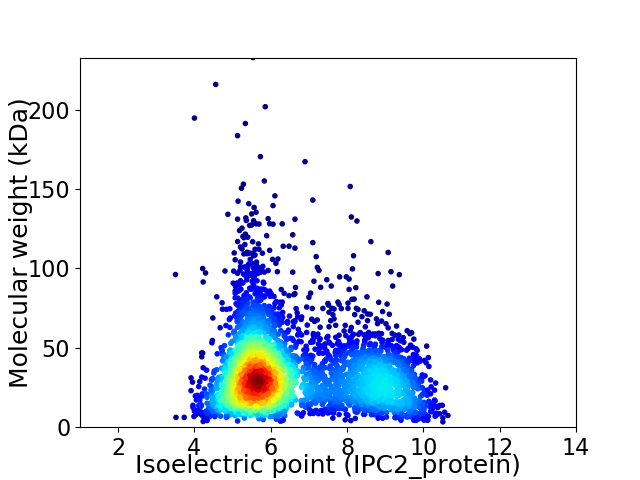
Virtual 2D-PAGE plot for 3981 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A1K2R6|A1K2R6_AZOSB RNA polymerase sigma-54 factor OS=Azoarcus sp. (strain BH72) OX=418699 GN=rpoN1 PE=3 SV=1
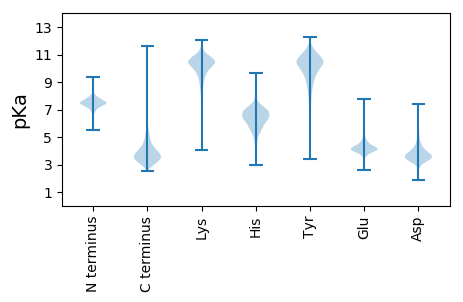
MM1 pKa = 7.95EE2 pKa = 4.6IRR4 pKa = 11.84KK5 pKa = 9.08VVLGCATACCAIASPSVSAALLPLPDD31 pKa = 3.73TVIDD35 pKa = 4.13GVVVSIQYY43 pKa = 11.25GDD45 pKa = 4.12FYY47 pKa = 11.33SYY49 pKa = 8.95STRR52 pKa = 11.84VLDD55 pKa = 4.31YY56 pKa = 10.13LQPDD60 pKa = 3.92AGWDD64 pKa = 3.42KK65 pKa = 11.4SAGTGTLDD73 pKa = 3.96VIITTRR79 pKa = 11.84SAGQSNSNGGLAPYY93 pKa = 9.36NIPDD97 pKa = 5.58PITNVNEE104 pKa = 4.11NPTSGSWGMGGTSATTMLVSDD125 pKa = 5.02LYY127 pKa = 10.97RR128 pKa = 11.84YY129 pKa = 9.67LQDD132 pKa = 3.8TFDD135 pKa = 3.17ATIPVFTFDD144 pKa = 4.14QNEE147 pKa = 4.4TGGNPSLFAMAKK159 pKa = 10.1VEE161 pKa = 4.91IFDD164 pKa = 3.99KK165 pKa = 11.06DD166 pKa = 3.63GNLLEE171 pKa = 4.92DD172 pKa = 4.67WILGDD177 pKa = 3.54GVNPVEE183 pKa = 5.01APGTVCVDD191 pKa = 3.65PDD193 pKa = 4.29SLPLKK198 pKa = 10.21CFSNNVGSGAFDD210 pKa = 4.19YY211 pKa = 11.11ILFAPTMDD219 pKa = 3.33LTNYY223 pKa = 9.18NAEE226 pKa = 3.99GNIFKK231 pKa = 10.88VSWTFSSVDD240 pKa = 3.58DD241 pKa = 4.51GGEE244 pKa = 3.87EE245 pKa = 3.83ATLTGRR251 pKa = 11.84FTGSICVEE259 pKa = 4.13NPSAPQCQVIPEE271 pKa = 4.49PGALALIGGGLLALFALRR289 pKa = 11.84RR290 pKa = 11.84RR291 pKa = 11.84WGAA294 pKa = 2.95
MM1 pKa = 7.95EE2 pKa = 4.6IRR4 pKa = 11.84KK5 pKa = 9.08VVLGCATACCAIASPSVSAALLPLPDD31 pKa = 3.73TVIDD35 pKa = 4.13GVVVSIQYY43 pKa = 11.25GDD45 pKa = 4.12FYY47 pKa = 11.33SYY49 pKa = 8.95STRR52 pKa = 11.84VLDD55 pKa = 4.31YY56 pKa = 10.13LQPDD60 pKa = 3.92AGWDD64 pKa = 3.42KK65 pKa = 11.4SAGTGTLDD73 pKa = 3.96VIITTRR79 pKa = 11.84SAGQSNSNGGLAPYY93 pKa = 9.36NIPDD97 pKa = 5.58PITNVNEE104 pKa = 4.11NPTSGSWGMGGTSATTMLVSDD125 pKa = 5.02LYY127 pKa = 10.97RR128 pKa = 11.84YY129 pKa = 9.67LQDD132 pKa = 3.8TFDD135 pKa = 3.17ATIPVFTFDD144 pKa = 4.14QNEE147 pKa = 4.4TGGNPSLFAMAKK159 pKa = 10.1VEE161 pKa = 4.91IFDD164 pKa = 3.99KK165 pKa = 11.06DD166 pKa = 3.63GNLLEE171 pKa = 4.92DD172 pKa = 4.67WILGDD177 pKa = 3.54GVNPVEE183 pKa = 5.01APGTVCVDD191 pKa = 3.65PDD193 pKa = 4.29SLPLKK198 pKa = 10.21CFSNNVGSGAFDD210 pKa = 4.19YY211 pKa = 11.11ILFAPTMDD219 pKa = 3.33LTNYY223 pKa = 9.18NAEE226 pKa = 3.99GNIFKK231 pKa = 10.88VSWTFSSVDD240 pKa = 3.58DD241 pKa = 4.51GGEE244 pKa = 3.87EE245 pKa = 3.83ATLTGRR251 pKa = 11.84FTGSICVEE259 pKa = 4.13NPSAPQCQVIPEE271 pKa = 4.49PGALALIGGGLLALFALRR289 pKa = 11.84RR290 pKa = 11.84RR291 pKa = 11.84WGAA294 pKa = 2.95
Molecular weight: 31.05 kDa
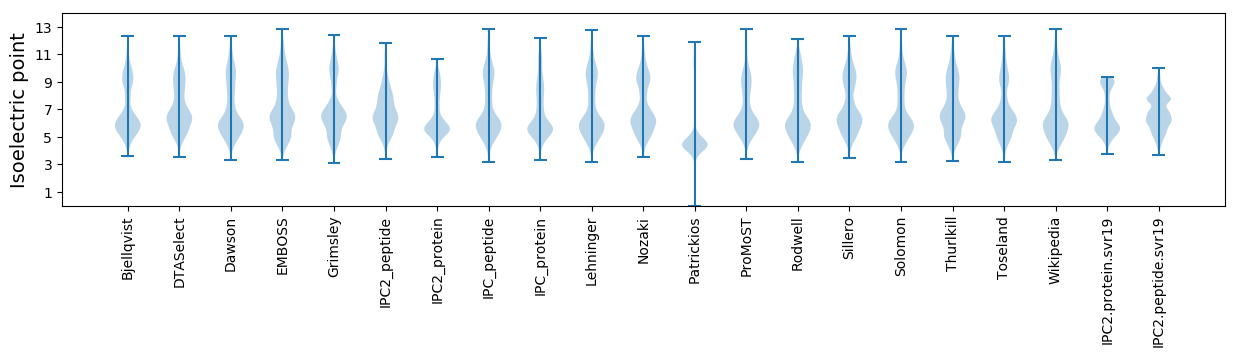
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A1K2C6|A1K2C6_AZOSB Conserved hypothetical membrane protein OS=Azoarcus sp. (strain BH72) OX=418699 GN=azo0364 PE=4 SV=1
MM1 pKa = 7.33MSRR4 pKa = 11.84IKK6 pKa = 10.91GRR8 pKa = 11.84DD9 pKa = 3.19TGPEE13 pKa = 3.35LSLRR17 pKa = 11.84RR18 pKa = 11.84NIWALGLRR26 pKa = 11.84YY27 pKa = 9.58RR28 pKa = 11.84LQYY31 pKa = 10.47RR32 pKa = 11.84IGRR35 pKa = 11.84TRR37 pKa = 11.84PDD39 pKa = 3.19MVFVRR44 pKa = 11.84ARR46 pKa = 11.84LAVFVDD52 pKa = 4.01GCFWHH57 pKa = 7.06GCPQHH62 pKa = 6.05STMPKK67 pKa = 8.63NNRR70 pKa = 11.84DD71 pKa = 2.89FWEE74 pKa = 4.16RR75 pKa = 11.84KK76 pKa = 8.1LRR78 pKa = 11.84RR79 pKa = 11.84NRR81 pKa = 11.84EE82 pKa = 3.3RR83 pKa = 11.84DD84 pKa = 3.25AEE86 pKa = 4.28NTHH89 pKa = 5.76SLEE92 pKa = 4.44AEE94 pKa = 4.14GWRR97 pKa = 11.84VLRR100 pKa = 11.84LWEE103 pKa = 4.35HH104 pKa = 7.28EE105 pKa = 4.25IQASPADD112 pKa = 3.82CARR115 pKa = 11.84RR116 pKa = 11.84IAVMLGKK123 pKa = 10.31AEE125 pKa = 3.98EE126 pKa = 4.47SKK128 pKa = 11.31AA129 pKa = 3.53
MM1 pKa = 7.33MSRR4 pKa = 11.84IKK6 pKa = 10.91GRR8 pKa = 11.84DD9 pKa = 3.19TGPEE13 pKa = 3.35LSLRR17 pKa = 11.84RR18 pKa = 11.84NIWALGLRR26 pKa = 11.84YY27 pKa = 9.58RR28 pKa = 11.84LQYY31 pKa = 10.47RR32 pKa = 11.84IGRR35 pKa = 11.84TRR37 pKa = 11.84PDD39 pKa = 3.19MVFVRR44 pKa = 11.84ARR46 pKa = 11.84LAVFVDD52 pKa = 4.01GCFWHH57 pKa = 7.06GCPQHH62 pKa = 6.05STMPKK67 pKa = 8.63NNRR70 pKa = 11.84DD71 pKa = 2.89FWEE74 pKa = 4.16RR75 pKa = 11.84KK76 pKa = 8.1LRR78 pKa = 11.84RR79 pKa = 11.84NRR81 pKa = 11.84EE82 pKa = 3.3RR83 pKa = 11.84DD84 pKa = 3.25AEE86 pKa = 4.28NTHH89 pKa = 5.76SLEE92 pKa = 4.44AEE94 pKa = 4.14GWRR97 pKa = 11.84VLRR100 pKa = 11.84LWEE103 pKa = 4.35HH104 pKa = 7.28EE105 pKa = 4.25IQASPADD112 pKa = 3.82CARR115 pKa = 11.84RR116 pKa = 11.84IAVMLGKK123 pKa = 10.31AEE125 pKa = 3.98EE126 pKa = 4.47SKK128 pKa = 11.31AA129 pKa = 3.53
Molecular weight: 15.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1326302 |
31 |
2109 |
333.2 |
36.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.574 ± 0.066 | 0.97 ± 0.015 |
5.492 ± 0.027 | 5.784 ± 0.036 |
3.547 ± 0.025 | 8.511 ± 0.029 |
2.272 ± 0.019 | 4.266 ± 0.026 |
2.737 ± 0.031 | 11.048 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.185 ± 0.018 | 2.412 ± 0.022 |
5.339 ± 0.031 | 3.326 ± 0.023 |
7.788 ± 0.04 | 4.759 ± 0.027 |
4.735 ± 0.032 | 7.603 ± 0.034 |
1.387 ± 0.014 | 2.265 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
