
Podoviridae sp. ctbd591
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
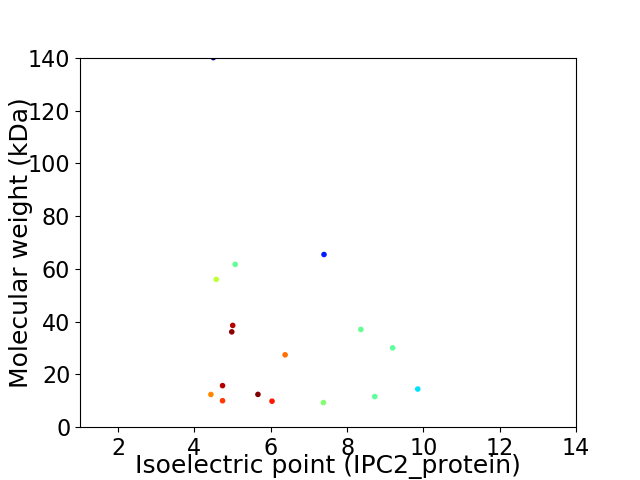
Virtual 2D-PAGE plot for 17 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345BNP6|A0A345BNP6_9CAUD DNA-directed DNA polymerase OS=Podoviridae sp. ctbd591 OX=2675445 PE=4 SV=1
MM1 pKa = 7.75SYY3 pKa = 11.28LDD5 pKa = 4.77DD6 pKa = 4.78GCHH9 pKa = 4.3NTYY12 pKa = 10.61FNGVPDD18 pKa = 4.26GCNKK22 pKa = 9.54KK23 pKa = 9.52HH24 pKa = 6.46CNNHH28 pKa = 4.37GCYY31 pKa = 9.93DD32 pKa = 4.01RR33 pKa = 11.84PPNCEE38 pKa = 3.79PNKK41 pKa = 10.36LDD43 pKa = 3.73SCEE46 pKa = 4.31TGCLPPPIKK55 pKa = 10.06PMCIIPGMNSQQQMSVIVRR74 pKa = 11.84KK75 pKa = 8.15TNEE78 pKa = 4.24CIEE81 pKa = 4.16RR82 pKa = 11.84WNTIQSNCYY91 pKa = 8.75EE92 pKa = 4.34ALNKK96 pKa = 10.35CVGAAVSNGVYY107 pKa = 10.17YY108 pKa = 10.69DD109 pKa = 3.79CEE111 pKa = 4.1EE112 pKa = 3.88VRR114 pKa = 11.84LQNGYY119 pKa = 9.6SEE121 pKa = 5.38NDD123 pKa = 3.08NCPYY127 pKa = 10.24KK128 pKa = 10.86YY129 pKa = 10.93VVIKK133 pKa = 10.11ATDD136 pKa = 3.3KK137 pKa = 10.62KK138 pKa = 10.76GCPIKK143 pKa = 11.04VKK145 pKa = 10.89LKK147 pKa = 8.88TAYY150 pKa = 10.24GNSGNNGARR159 pKa = 11.84QSMQDD164 pKa = 2.64ASFIYY169 pKa = 10.18SANTIISAINPVSGRR184 pKa = 11.84WSGPALIDD192 pKa = 3.87GCPMEE197 pKa = 4.58YY198 pKa = 9.61IKK200 pKa = 10.94PEE202 pKa = 4.23TIVDD206 pKa = 3.57GDD208 pKa = 3.68VLYY211 pKa = 11.09GFNKK215 pKa = 9.07GGAMLCFGPNTEE227 pKa = 4.73LEE229 pKa = 4.25TLKK232 pKa = 9.89MCGMVDD238 pKa = 3.81VIGMGIQILKK248 pKa = 10.23DD249 pKa = 3.67GNVTAAAEE257 pKa = 4.18SLTTGASIQAIGYY270 pKa = 9.2KK271 pKa = 9.81SSCGEE276 pKa = 3.96KK277 pKa = 10.68VFFSCGKK284 pKa = 9.02EE285 pKa = 3.9NSPGMQGINVANVLKK300 pKa = 11.27NMGCTTAVITCLQTMYY316 pKa = 10.89PDD318 pKa = 3.27ITGTTYY324 pKa = 10.68RR325 pKa = 11.84GIEE328 pKa = 4.12NAVTPASDD336 pKa = 5.62AIQQKK341 pKa = 8.88TGLTGGVMFLGRR353 pKa = 11.84LLNPPLEE360 pKa = 4.09WQIPYY365 pKa = 9.96NQAFWVVSKK374 pKa = 10.83KK375 pKa = 10.19PSNGFRR381 pKa = 11.84NDD383 pKa = 2.88FTTEE387 pKa = 4.01VADD390 pKa = 3.88LTQSLGGSVNNMASFANVIDD410 pKa = 3.75NMQIDD415 pKa = 4.39VLEE418 pKa = 4.3VTQKK422 pKa = 10.15IEE424 pKa = 4.35KK425 pKa = 10.02IEE427 pKa = 4.21SDD429 pKa = 3.4VSNISEE435 pKa = 5.13SITQLEE441 pKa = 4.34TRR443 pKa = 11.84VNAIEE448 pKa = 4.24PKK450 pKa = 9.24LTNLEE455 pKa = 4.2SDD457 pKa = 3.67VEE459 pKa = 4.34TLKK462 pKa = 11.0NCCNTVQEE470 pKa = 4.4YY471 pKa = 8.72MVTEE475 pKa = 4.09TEE477 pKa = 3.97EE478 pKa = 4.43RR479 pKa = 11.84KK480 pKa = 10.15QSDD483 pKa = 3.42TALGDD488 pKa = 4.04RR489 pKa = 11.84ITALQIAIQEE499 pKa = 4.3EE500 pKa = 4.56ATTRR504 pKa = 11.84NDD506 pKa = 3.27ADD508 pKa = 3.32NRR510 pKa = 11.84LIEE513 pKa = 4.94AIQNEE518 pKa = 4.14ASARR522 pKa = 11.84ATNDD526 pKa = 2.79AKK528 pKa = 11.11LDD530 pKa = 3.67TKK532 pKa = 10.6IDD534 pKa = 3.77SEE536 pKa = 4.61VAKK539 pKa = 10.62LEE541 pKa = 4.24SAIQDD546 pKa = 3.57IEE548 pKa = 4.38VAAYY552 pKa = 7.31TAGEE556 pKa = 4.2GIIINNRR563 pKa = 11.84VISVNKK569 pKa = 10.2DD570 pKa = 3.25DD571 pKa = 5.79FPDD574 pKa = 3.6NEE576 pKa = 4.89KK577 pKa = 10.47ISEE580 pKa = 4.26LEE582 pKa = 4.02NKK584 pKa = 9.39VNALPNYY591 pKa = 9.3VAGEE595 pKa = 4.41HH596 pKa = 6.52IVFTQSGGNTSISADD611 pKa = 3.43LTDD614 pKa = 4.15LSEE617 pKa = 4.63EE618 pKa = 4.06IEE620 pKa = 4.15NTKK623 pKa = 10.27TIVQEE628 pKa = 3.99NTSKK632 pKa = 10.55IEE634 pKa = 3.88KK635 pKa = 10.48AEE637 pKa = 3.87GDD639 pKa = 3.61IEE641 pKa = 4.52NNASSITTIEE651 pKa = 4.2NEE653 pKa = 3.9VNTIKK658 pKa = 11.06GDD660 pKa = 3.48ISQLQSDD667 pKa = 4.2VKK669 pKa = 10.24EE670 pKa = 4.29ANISAGNGIEE680 pKa = 4.19INTVDD685 pKa = 3.91DD686 pKa = 4.02SKK688 pKa = 11.24EE689 pKa = 3.68ISVKK693 pKa = 9.92IDD695 pKa = 3.2STLSFQPDD703 pKa = 3.42GALHH707 pKa = 6.42VDD709 pKa = 4.3LPAPEE714 pKa = 4.37TGEE717 pKa = 4.2TTRR720 pKa = 11.84AGYY723 pKa = 10.33GIIISKK729 pKa = 10.64NEE731 pKa = 4.56DD732 pKa = 2.93IATIAVNQQLVASTADD748 pKa = 3.1IQNIKK753 pKa = 10.54SGTTPLPYY761 pKa = 10.52VKK763 pKa = 10.55VSGDD767 pKa = 2.89IMTGEE772 pKa = 4.88LIGTTFKK779 pKa = 11.43SNVEE783 pKa = 3.71MPIRR787 pKa = 11.84DD788 pKa = 3.58NNEE791 pKa = 3.25NHH793 pKa = 6.31SAFGVDD799 pKa = 4.3VEE801 pKa = 4.88DD802 pKa = 4.43NKK804 pKa = 11.09LKK806 pKa = 10.9YY807 pKa = 10.54GGVGEE812 pKa = 3.94NTVFNNRR819 pKa = 11.84VKK821 pKa = 9.55ITNVDD826 pKa = 3.79TPSGDD831 pKa = 3.32TDD833 pKa = 3.83ATPKK837 pKa = 10.78DD838 pKa = 3.83YY839 pKa = 11.63VDD841 pKa = 4.02NKK843 pKa = 10.6DD844 pKa = 3.59DD845 pKa = 4.45AVISMTNTAIKK856 pKa = 10.21SAEE859 pKa = 3.78DD860 pKa = 3.9RR861 pKa = 11.84IEE863 pKa = 4.31GEE865 pKa = 3.62IAANTTEE872 pKa = 4.07INEE875 pKa = 4.53KK876 pKa = 10.26IEE878 pKa = 4.15STKK881 pKa = 11.07NEE883 pKa = 4.06LQNNINVIDD892 pKa = 3.92EE893 pKa = 4.63AQTEE897 pKa = 4.2LQEE900 pKa = 4.14QFSKK904 pKa = 11.47LEE906 pKa = 4.69DD907 pKa = 3.98GSVSLPYY914 pKa = 10.3VKK916 pKa = 10.53KK917 pKa = 10.87SGDD920 pKa = 3.67SMSGPLSLVDD930 pKa = 3.17STGAVLGQLSLTEE943 pKa = 4.22NGLMILGGPAATGAHH958 pKa = 4.89LTITNEE964 pKa = 4.15SVSIDD969 pKa = 3.5KK970 pKa = 8.0PTHH973 pKa = 5.51VNGTVSANNATKK985 pKa = 10.67DD986 pKa = 3.65NEE988 pKa = 4.44LVTLEE993 pKa = 4.08QLNAKK998 pKa = 9.67PSGISQTDD1006 pKa = 2.61ADD1008 pKa = 3.82ARR1010 pKa = 11.84YY1011 pKa = 8.4IASSNGVSHH1020 pKa = 6.41TPVYY1024 pKa = 10.47FDD1026 pKa = 5.52AEE1028 pKa = 4.15EE1029 pKa = 4.49DD1030 pKa = 3.75LNGWVHH1036 pKa = 7.5IYY1038 pKa = 10.81RR1039 pKa = 11.84LTKK1042 pKa = 10.87GNGLQSAINHH1052 pKa = 6.24NGIQYY1057 pKa = 9.94NSKK1060 pKa = 10.82DD1061 pKa = 3.08ITTTIDD1067 pKa = 3.37NPIPLQIGNPTLPSHH1082 pKa = 7.08AANKK1086 pKa = 10.31SYY1088 pKa = 11.32VDD1090 pKa = 3.81AVASSVGDD1098 pKa = 3.64GKK1100 pKa = 10.8FLPLTGGTMSGPIYY1114 pKa = 9.55ITEE1117 pKa = 4.2HH1118 pKa = 6.39NAFQSPGRR1126 pKa = 11.84QVPTFFSIDD1135 pKa = 3.47ARR1137 pKa = 11.84SGLYY1141 pKa = 10.45FSDD1144 pKa = 5.04DD1145 pKa = 3.4NNLEE1149 pKa = 4.13QNPVRR1154 pKa = 11.84VSGVGDD1160 pKa = 3.41PTIDD1164 pKa = 2.93IDD1166 pKa = 4.14AANKK1170 pKa = 10.19RR1171 pKa = 11.84YY1172 pKa = 10.02VDD1174 pKa = 3.8SKK1176 pKa = 10.12TDD1178 pKa = 3.33GLGYY1182 pKa = 9.78PVVKK1186 pKa = 10.4SIQEE1190 pKa = 4.03LVSAAVAGATKK1201 pKa = 9.6TISGGMVLTWGRR1213 pKa = 11.84LLYY1216 pKa = 10.29ISITFRR1222 pKa = 11.84VDD1224 pKa = 2.88EE1225 pKa = 4.3TLSNGTRR1232 pKa = 11.84LPFYY1236 pKa = 10.65VQLPSQGFYY1245 pKa = 10.37QGNPLVSATPGVGVGTTPYY1264 pKa = 10.95CNITVGTPLYY1274 pKa = 10.16SGSNMFVQGCIFGTDD1289 pKa = 2.93TYY1291 pKa = 11.07ALQNITVTMLGCAA1304 pKa = 3.83
MM1 pKa = 7.75SYY3 pKa = 11.28LDD5 pKa = 4.77DD6 pKa = 4.78GCHH9 pKa = 4.3NTYY12 pKa = 10.61FNGVPDD18 pKa = 4.26GCNKK22 pKa = 9.54KK23 pKa = 9.52HH24 pKa = 6.46CNNHH28 pKa = 4.37GCYY31 pKa = 9.93DD32 pKa = 4.01RR33 pKa = 11.84PPNCEE38 pKa = 3.79PNKK41 pKa = 10.36LDD43 pKa = 3.73SCEE46 pKa = 4.31TGCLPPPIKK55 pKa = 10.06PMCIIPGMNSQQQMSVIVRR74 pKa = 11.84KK75 pKa = 8.15TNEE78 pKa = 4.24CIEE81 pKa = 4.16RR82 pKa = 11.84WNTIQSNCYY91 pKa = 8.75EE92 pKa = 4.34ALNKK96 pKa = 10.35CVGAAVSNGVYY107 pKa = 10.17YY108 pKa = 10.69DD109 pKa = 3.79CEE111 pKa = 4.1EE112 pKa = 3.88VRR114 pKa = 11.84LQNGYY119 pKa = 9.6SEE121 pKa = 5.38NDD123 pKa = 3.08NCPYY127 pKa = 10.24KK128 pKa = 10.86YY129 pKa = 10.93VVIKK133 pKa = 10.11ATDD136 pKa = 3.3KK137 pKa = 10.62KK138 pKa = 10.76GCPIKK143 pKa = 11.04VKK145 pKa = 10.89LKK147 pKa = 8.88TAYY150 pKa = 10.24GNSGNNGARR159 pKa = 11.84QSMQDD164 pKa = 2.64ASFIYY169 pKa = 10.18SANTIISAINPVSGRR184 pKa = 11.84WSGPALIDD192 pKa = 3.87GCPMEE197 pKa = 4.58YY198 pKa = 9.61IKK200 pKa = 10.94PEE202 pKa = 4.23TIVDD206 pKa = 3.57GDD208 pKa = 3.68VLYY211 pKa = 11.09GFNKK215 pKa = 9.07GGAMLCFGPNTEE227 pKa = 4.73LEE229 pKa = 4.25TLKK232 pKa = 9.89MCGMVDD238 pKa = 3.81VIGMGIQILKK248 pKa = 10.23DD249 pKa = 3.67GNVTAAAEE257 pKa = 4.18SLTTGASIQAIGYY270 pKa = 9.2KK271 pKa = 9.81SSCGEE276 pKa = 3.96KK277 pKa = 10.68VFFSCGKK284 pKa = 9.02EE285 pKa = 3.9NSPGMQGINVANVLKK300 pKa = 11.27NMGCTTAVITCLQTMYY316 pKa = 10.89PDD318 pKa = 3.27ITGTTYY324 pKa = 10.68RR325 pKa = 11.84GIEE328 pKa = 4.12NAVTPASDD336 pKa = 5.62AIQQKK341 pKa = 8.88TGLTGGVMFLGRR353 pKa = 11.84LLNPPLEE360 pKa = 4.09WQIPYY365 pKa = 9.96NQAFWVVSKK374 pKa = 10.83KK375 pKa = 10.19PSNGFRR381 pKa = 11.84NDD383 pKa = 2.88FTTEE387 pKa = 4.01VADD390 pKa = 3.88LTQSLGGSVNNMASFANVIDD410 pKa = 3.75NMQIDD415 pKa = 4.39VLEE418 pKa = 4.3VTQKK422 pKa = 10.15IEE424 pKa = 4.35KK425 pKa = 10.02IEE427 pKa = 4.21SDD429 pKa = 3.4VSNISEE435 pKa = 5.13SITQLEE441 pKa = 4.34TRR443 pKa = 11.84VNAIEE448 pKa = 4.24PKK450 pKa = 9.24LTNLEE455 pKa = 4.2SDD457 pKa = 3.67VEE459 pKa = 4.34TLKK462 pKa = 11.0NCCNTVQEE470 pKa = 4.4YY471 pKa = 8.72MVTEE475 pKa = 4.09TEE477 pKa = 3.97EE478 pKa = 4.43RR479 pKa = 11.84KK480 pKa = 10.15QSDD483 pKa = 3.42TALGDD488 pKa = 4.04RR489 pKa = 11.84ITALQIAIQEE499 pKa = 4.3EE500 pKa = 4.56ATTRR504 pKa = 11.84NDD506 pKa = 3.27ADD508 pKa = 3.32NRR510 pKa = 11.84LIEE513 pKa = 4.94AIQNEE518 pKa = 4.14ASARR522 pKa = 11.84ATNDD526 pKa = 2.79AKK528 pKa = 11.11LDD530 pKa = 3.67TKK532 pKa = 10.6IDD534 pKa = 3.77SEE536 pKa = 4.61VAKK539 pKa = 10.62LEE541 pKa = 4.24SAIQDD546 pKa = 3.57IEE548 pKa = 4.38VAAYY552 pKa = 7.31TAGEE556 pKa = 4.2GIIINNRR563 pKa = 11.84VISVNKK569 pKa = 10.2DD570 pKa = 3.25DD571 pKa = 5.79FPDD574 pKa = 3.6NEE576 pKa = 4.89KK577 pKa = 10.47ISEE580 pKa = 4.26LEE582 pKa = 4.02NKK584 pKa = 9.39VNALPNYY591 pKa = 9.3VAGEE595 pKa = 4.41HH596 pKa = 6.52IVFTQSGGNTSISADD611 pKa = 3.43LTDD614 pKa = 4.15LSEE617 pKa = 4.63EE618 pKa = 4.06IEE620 pKa = 4.15NTKK623 pKa = 10.27TIVQEE628 pKa = 3.99NTSKK632 pKa = 10.55IEE634 pKa = 3.88KK635 pKa = 10.48AEE637 pKa = 3.87GDD639 pKa = 3.61IEE641 pKa = 4.52NNASSITTIEE651 pKa = 4.2NEE653 pKa = 3.9VNTIKK658 pKa = 11.06GDD660 pKa = 3.48ISQLQSDD667 pKa = 4.2VKK669 pKa = 10.24EE670 pKa = 4.29ANISAGNGIEE680 pKa = 4.19INTVDD685 pKa = 3.91DD686 pKa = 4.02SKK688 pKa = 11.24EE689 pKa = 3.68ISVKK693 pKa = 9.92IDD695 pKa = 3.2STLSFQPDD703 pKa = 3.42GALHH707 pKa = 6.42VDD709 pKa = 4.3LPAPEE714 pKa = 4.37TGEE717 pKa = 4.2TTRR720 pKa = 11.84AGYY723 pKa = 10.33GIIISKK729 pKa = 10.64NEE731 pKa = 4.56DD732 pKa = 2.93IATIAVNQQLVASTADD748 pKa = 3.1IQNIKK753 pKa = 10.54SGTTPLPYY761 pKa = 10.52VKK763 pKa = 10.55VSGDD767 pKa = 2.89IMTGEE772 pKa = 4.88LIGTTFKK779 pKa = 11.43SNVEE783 pKa = 3.71MPIRR787 pKa = 11.84DD788 pKa = 3.58NNEE791 pKa = 3.25NHH793 pKa = 6.31SAFGVDD799 pKa = 4.3VEE801 pKa = 4.88DD802 pKa = 4.43NKK804 pKa = 11.09LKK806 pKa = 10.9YY807 pKa = 10.54GGVGEE812 pKa = 3.94NTVFNNRR819 pKa = 11.84VKK821 pKa = 9.55ITNVDD826 pKa = 3.79TPSGDD831 pKa = 3.32TDD833 pKa = 3.83ATPKK837 pKa = 10.78DD838 pKa = 3.83YY839 pKa = 11.63VDD841 pKa = 4.02NKK843 pKa = 10.6DD844 pKa = 3.59DD845 pKa = 4.45AVISMTNTAIKK856 pKa = 10.21SAEE859 pKa = 3.78DD860 pKa = 3.9RR861 pKa = 11.84IEE863 pKa = 4.31GEE865 pKa = 3.62IAANTTEE872 pKa = 4.07INEE875 pKa = 4.53KK876 pKa = 10.26IEE878 pKa = 4.15STKK881 pKa = 11.07NEE883 pKa = 4.06LQNNINVIDD892 pKa = 3.92EE893 pKa = 4.63AQTEE897 pKa = 4.2LQEE900 pKa = 4.14QFSKK904 pKa = 11.47LEE906 pKa = 4.69DD907 pKa = 3.98GSVSLPYY914 pKa = 10.3VKK916 pKa = 10.53KK917 pKa = 10.87SGDD920 pKa = 3.67SMSGPLSLVDD930 pKa = 3.17STGAVLGQLSLTEE943 pKa = 4.22NGLMILGGPAATGAHH958 pKa = 4.89LTITNEE964 pKa = 4.15SVSIDD969 pKa = 3.5KK970 pKa = 8.0PTHH973 pKa = 5.51VNGTVSANNATKK985 pKa = 10.67DD986 pKa = 3.65NEE988 pKa = 4.44LVTLEE993 pKa = 4.08QLNAKK998 pKa = 9.67PSGISQTDD1006 pKa = 2.61ADD1008 pKa = 3.82ARR1010 pKa = 11.84YY1011 pKa = 8.4IASSNGVSHH1020 pKa = 6.41TPVYY1024 pKa = 10.47FDD1026 pKa = 5.52AEE1028 pKa = 4.15EE1029 pKa = 4.49DD1030 pKa = 3.75LNGWVHH1036 pKa = 7.5IYY1038 pKa = 10.81RR1039 pKa = 11.84LTKK1042 pKa = 10.87GNGLQSAINHH1052 pKa = 6.24NGIQYY1057 pKa = 9.94NSKK1060 pKa = 10.82DD1061 pKa = 3.08ITTTIDD1067 pKa = 3.37NPIPLQIGNPTLPSHH1082 pKa = 7.08AANKK1086 pKa = 10.31SYY1088 pKa = 11.32VDD1090 pKa = 3.81AVASSVGDD1098 pKa = 3.64GKK1100 pKa = 10.8FLPLTGGTMSGPIYY1114 pKa = 9.55ITEE1117 pKa = 4.2HH1118 pKa = 6.39NAFQSPGRR1126 pKa = 11.84QVPTFFSIDD1135 pKa = 3.47ARR1137 pKa = 11.84SGLYY1141 pKa = 10.45FSDD1144 pKa = 5.04DD1145 pKa = 3.4NNLEE1149 pKa = 4.13QNPVRR1154 pKa = 11.84VSGVGDD1160 pKa = 3.41PTIDD1164 pKa = 2.93IDD1166 pKa = 4.14AANKK1170 pKa = 10.19RR1171 pKa = 11.84YY1172 pKa = 10.02VDD1174 pKa = 3.8SKK1176 pKa = 10.12TDD1178 pKa = 3.33GLGYY1182 pKa = 9.78PVVKK1186 pKa = 10.4SIQEE1190 pKa = 4.03LVSAAVAGATKK1201 pKa = 9.6TISGGMVLTWGRR1213 pKa = 11.84LLYY1216 pKa = 10.29ISITFRR1222 pKa = 11.84VDD1224 pKa = 2.88EE1225 pKa = 4.3TLSNGTRR1232 pKa = 11.84LPFYY1236 pKa = 10.65VQLPSQGFYY1245 pKa = 10.37QGNPLVSATPGVGVGTTPYY1264 pKa = 10.95CNITVGTPLYY1274 pKa = 10.16SGSNMFVQGCIFGTDD1289 pKa = 2.93TYY1291 pKa = 11.07ALQNITVTMLGCAA1304 pKa = 3.83
Molecular weight: 140.15 kDa
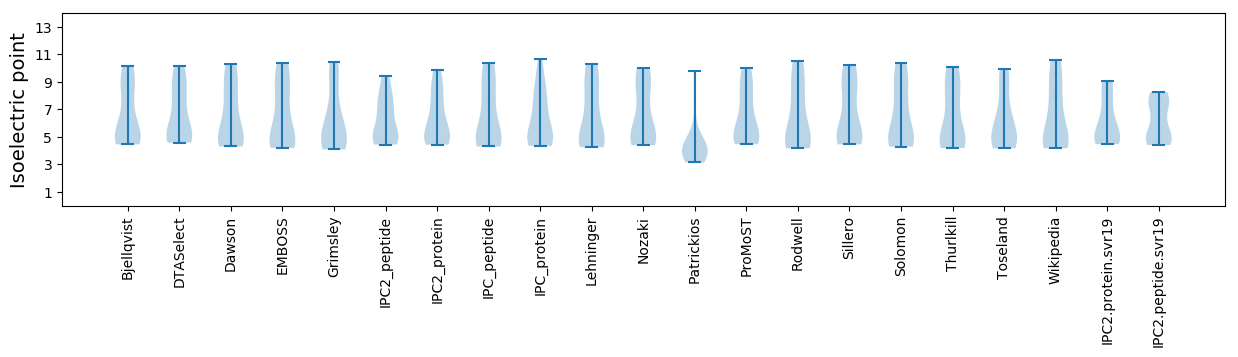
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345BNN9|A0A345BNN9_9CAUD Uncharacterized protein OS=Podoviridae sp. ctbd591 OX=2675445 PE=4 SV=1
MM1 pKa = 7.71SKK3 pKa = 10.56RR4 pKa = 11.84KK5 pKa = 9.2NSKK8 pKa = 8.72NKK10 pKa = 10.45GYY12 pKa = 8.52ITLPGGRR19 pKa = 11.84VRR21 pKa = 11.84RR22 pKa = 11.84STIDD26 pKa = 3.13NYY28 pKa = 11.0RR29 pKa = 11.84KK30 pKa = 9.59AVKK33 pKa = 10.11NYY35 pKa = 9.21NEE37 pKa = 4.54RR38 pKa = 11.84IRR40 pKa = 11.84YY41 pKa = 8.1AKK43 pKa = 10.26RR44 pKa = 11.84KK45 pKa = 8.65FGNYY49 pKa = 9.73DD50 pKa = 3.71YY51 pKa = 11.1LPKK54 pKa = 10.11PIKK57 pKa = 10.51ASEE60 pKa = 4.11TLKK63 pKa = 11.09GFTDD67 pKa = 4.45GKK69 pKa = 9.87SIRR72 pKa = 11.84DD73 pKa = 4.46FISDD77 pKa = 3.26INKK80 pKa = 9.56INISNLTPVEE90 pKa = 4.26TKK92 pKa = 10.69AGVITKK98 pKa = 10.7GEE100 pKa = 3.97LKK102 pKa = 10.05MIKK105 pKa = 8.88TRR107 pKa = 11.84IKK109 pKa = 10.49RR110 pKa = 11.84EE111 pKa = 3.79NKK113 pKa = 9.39RR114 pKa = 11.84RR115 pKa = 11.84AQVKK119 pKa = 10.32KK120 pKa = 10.8LVDD123 pKa = 3.39KK124 pKa = 11.14QEE126 pKa = 3.88NEE128 pKa = 3.26IGYY131 pKa = 9.93FKK133 pKa = 10.58TDD135 pKa = 2.79NDD137 pKa = 4.79KK138 pKa = 11.12ILMPIDD144 pKa = 3.09IEE146 pKa = 4.35KK147 pKa = 10.82YY148 pKa = 10.17NNLEE152 pKa = 3.86SLRR155 pKa = 11.84RR156 pKa = 11.84LSDD159 pKa = 4.25RR160 pKa = 11.84ISVDD164 pKa = 3.14YY165 pKa = 10.74RR166 pKa = 11.84NKK168 pKa = 10.64LAFDD172 pKa = 3.13WKK174 pKa = 10.64DD175 pKa = 3.21RR176 pKa = 11.84YY177 pKa = 10.2VNQVTFVINQGLISGLSQDD196 pKa = 4.21DD197 pKa = 4.26EE198 pKa = 5.21IIVRR202 pKa = 11.84LEE204 pKa = 4.02TIKK207 pKa = 10.66RR208 pKa = 11.84WVLSLDD214 pKa = 3.07NTTDD218 pKa = 2.9ITRR221 pKa = 11.84AAYY224 pKa = 8.24AAPSMDD230 pKa = 2.61ITVIYY235 pKa = 9.95YY236 pKa = 9.64SEE238 pKa = 3.96ISGPIVSEE246 pKa = 4.72MYY248 pKa = 10.87NDD250 pKa = 3.28WNLFMDD256 pKa = 4.31SLL258 pKa = 4.26
MM1 pKa = 7.71SKK3 pKa = 10.56RR4 pKa = 11.84KK5 pKa = 9.2NSKK8 pKa = 8.72NKK10 pKa = 10.45GYY12 pKa = 8.52ITLPGGRR19 pKa = 11.84VRR21 pKa = 11.84RR22 pKa = 11.84STIDD26 pKa = 3.13NYY28 pKa = 11.0RR29 pKa = 11.84KK30 pKa = 9.59AVKK33 pKa = 10.11NYY35 pKa = 9.21NEE37 pKa = 4.54RR38 pKa = 11.84IRR40 pKa = 11.84YY41 pKa = 8.1AKK43 pKa = 10.26RR44 pKa = 11.84KK45 pKa = 8.65FGNYY49 pKa = 9.73DD50 pKa = 3.71YY51 pKa = 11.1LPKK54 pKa = 10.11PIKK57 pKa = 10.51ASEE60 pKa = 4.11TLKK63 pKa = 11.09GFTDD67 pKa = 4.45GKK69 pKa = 9.87SIRR72 pKa = 11.84DD73 pKa = 4.46FISDD77 pKa = 3.26INKK80 pKa = 9.56INISNLTPVEE90 pKa = 4.26TKK92 pKa = 10.69AGVITKK98 pKa = 10.7GEE100 pKa = 3.97LKK102 pKa = 10.05MIKK105 pKa = 8.88TRR107 pKa = 11.84IKK109 pKa = 10.49RR110 pKa = 11.84EE111 pKa = 3.79NKK113 pKa = 9.39RR114 pKa = 11.84RR115 pKa = 11.84AQVKK119 pKa = 10.32KK120 pKa = 10.8LVDD123 pKa = 3.39KK124 pKa = 11.14QEE126 pKa = 3.88NEE128 pKa = 3.26IGYY131 pKa = 9.93FKK133 pKa = 10.58TDD135 pKa = 2.79NDD137 pKa = 4.79KK138 pKa = 11.12ILMPIDD144 pKa = 3.09IEE146 pKa = 4.35KK147 pKa = 10.82YY148 pKa = 10.17NNLEE152 pKa = 3.86SLRR155 pKa = 11.84RR156 pKa = 11.84LSDD159 pKa = 4.25RR160 pKa = 11.84ISVDD164 pKa = 3.14YY165 pKa = 10.74RR166 pKa = 11.84NKK168 pKa = 10.64LAFDD172 pKa = 3.13WKK174 pKa = 10.64DD175 pKa = 3.21RR176 pKa = 11.84YY177 pKa = 10.2VNQVTFVINQGLISGLSQDD196 pKa = 4.21DD197 pKa = 4.26EE198 pKa = 5.21IIVRR202 pKa = 11.84LEE204 pKa = 4.02TIKK207 pKa = 10.66RR208 pKa = 11.84WVLSLDD214 pKa = 3.07NTTDD218 pKa = 2.9ITRR221 pKa = 11.84AAYY224 pKa = 8.24AAPSMDD230 pKa = 2.61ITVIYY235 pKa = 9.95YY236 pKa = 9.64SEE238 pKa = 3.96ISGPIVSEE246 pKa = 4.72MYY248 pKa = 10.87NDD250 pKa = 3.28WNLFMDD256 pKa = 4.31SLL258 pKa = 4.26
Molecular weight: 30.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5232 |
81 |
1304 |
307.8 |
34.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.657 ± 0.705 | 1.567 ± 0.306 |
5.944 ± 0.206 | 6.67 ± 0.526 |
4.434 ± 0.662 | 6.307 ± 0.659 |
1.281 ± 0.218 | 7.932 ± 0.547 |
6.384 ± 0.699 | 6.881 ± 0.619 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.255 ± 0.214 | 8.008 ± 0.651 |
3.345 ± 0.418 | 2.867 ± 0.481 |
4.014 ± 0.58 | 7.607 ± 0.407 |
7.148 ± 1.084 | 6.097 ± 0.466 |
1.013 ± 0.252 | 4.587 ± 0.526 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
