
Botrytis cinerea mitovirus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.56
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S4GAV3|A0A0S4GAV3_9VIRU RNA dependent RNA polymerase OS=Botrytis cinerea mitovirus 3 OX=1629666 GN=RdRp PE=4 SV=1
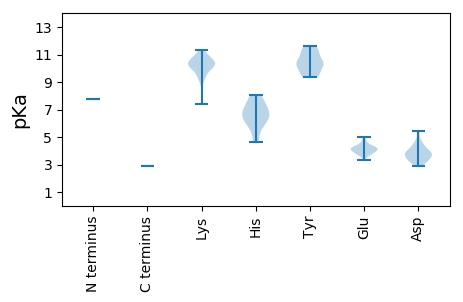
MM1 pKa = 7.76KK2 pKa = 9.36ITRR5 pKa = 11.84TYY7 pKa = 11.31KK8 pKa = 10.6NLNSLLKK15 pKa = 10.68SSIVKK20 pKa = 7.38TKK22 pKa = 10.85KK23 pKa = 9.28MFSIRR28 pKa = 11.84DD29 pKa = 3.78SISNSLSIPLRR40 pKa = 11.84QIQYY44 pKa = 10.93LSFGKK49 pKa = 10.05IRR51 pKa = 11.84GLANRR56 pKa = 11.84ALITRR61 pKa = 11.84DD62 pKa = 2.91FMTFVLKK69 pKa = 10.01MKK71 pKa = 9.87KK72 pKa = 9.35NHH74 pKa = 6.37GADD77 pKa = 4.26FTIKK81 pKa = 9.34WLKK84 pKa = 8.89CCYY87 pKa = 9.82VALQKK92 pKa = 11.13SLGKK96 pKa = 10.45DD97 pKa = 3.02NLEE100 pKa = 4.03SLRR103 pKa = 11.84DD104 pKa = 3.83LEE106 pKa = 4.96PNLPLPRR113 pKa = 11.84LINGIPAIISSNDD126 pKa = 2.94RR127 pKa = 11.84KK128 pKa = 10.47LIRR131 pKa = 11.84EE132 pKa = 3.89MDD134 pKa = 3.56SRR136 pKa = 11.84IIIYY140 pKa = 9.45WSSLFSLYY148 pKa = 10.18RR149 pKa = 11.84VLKK152 pKa = 10.38CSYY155 pKa = 9.35KK156 pKa = 10.85LKK158 pKa = 10.29ISSITDD164 pKa = 3.46PFKK167 pKa = 11.33GSINEE172 pKa = 4.11FNDD175 pKa = 3.92IIQGPVFSIFFDD187 pKa = 3.6RR188 pKa = 11.84LNGFPEE194 pKa = 4.29LVKK197 pKa = 10.73KK198 pKa = 11.08SNLAPSKK205 pKa = 10.62VRR207 pKa = 11.84LLRR210 pKa = 11.84SSSSSNNVSWHH221 pKa = 6.44GIITDD226 pKa = 3.07SWNISNSKK234 pKa = 9.86EE235 pKa = 3.69MSSNVSNYY243 pKa = 9.54LASIKK248 pKa = 10.72ALGWNTMWFNSKK260 pKa = 10.03LNEE263 pKa = 4.17MIEE266 pKa = 4.22LGDD269 pKa = 3.86RR270 pKa = 11.84LNQVGSLKK278 pKa = 9.81TKK280 pKa = 10.44KK281 pKa = 10.31SLSGQFGQFSLKK293 pKa = 10.46EE294 pKa = 3.73EE295 pKa = 4.19AAGKK299 pKa = 10.03LRR301 pKa = 11.84IFAIVDD307 pKa = 4.02SITQSLLSPLHH318 pKa = 7.02DD319 pKa = 4.61FMFDD323 pKa = 3.56LLKK326 pKa = 10.64KK327 pKa = 10.03IPNDD331 pKa = 3.47GTFDD335 pKa = 3.24QDD337 pKa = 3.69LSVKK341 pKa = 10.13RR342 pKa = 11.84SQVKK346 pKa = 10.2SLSSGKK352 pKa = 10.26AFSFDD357 pKa = 3.28LSAATDD363 pKa = 3.94RR364 pKa = 11.84LPVDD368 pKa = 3.99LTVKK372 pKa = 10.03ILSKK376 pKa = 10.49IFSDD380 pKa = 4.48EE381 pKa = 4.98FGTSWKK387 pKa = 10.27QLMVNRR393 pKa = 11.84DD394 pKa = 3.51FFFSLTNQKK403 pKa = 10.99DD404 pKa = 3.48YY405 pKa = 11.34GAPEE409 pKa = 3.99SLRR412 pKa = 11.84YY413 pKa = 9.59SVGQPMGALSSWPALALTHH432 pKa = 6.47HH433 pKa = 7.57WILQYY438 pKa = 10.7CSNILGRR445 pKa = 11.84TGWEE449 pKa = 3.65EE450 pKa = 3.73NYY452 pKa = 10.47EE453 pKa = 4.03ILGDD457 pKa = 3.9DD458 pKa = 4.07LVVFDD463 pKa = 5.41SALADD468 pKa = 3.65KK469 pKa = 10.42YY470 pKa = 11.59LEE472 pKa = 3.89IAKK475 pKa = 10.24ILGVEE480 pKa = 3.95INLTKK485 pKa = 10.57SISSHH490 pKa = 5.94DD491 pKa = 3.43RR492 pKa = 11.84PVFEE496 pKa = 4.56FAKK499 pKa = 9.26RR500 pKa = 11.84TCFGNSDD507 pKa = 4.53LSPISIKK514 pKa = 10.42QLLSNDD520 pKa = 3.48QLSEE524 pKa = 3.63RR525 pKa = 11.84TMNVVSFLKK534 pKa = 10.55RR535 pKa = 11.84GLLVSRR541 pKa = 11.84SHH543 pKa = 8.04IGILLSKK550 pKa = 10.18FGSWQILKK558 pKa = 10.26NKK560 pKa = 9.58KK561 pKa = 9.56LSKK564 pKa = 9.81TPLLAILGILNSMNLISHH582 pKa = 6.94RR583 pKa = 11.84WLTEE587 pKa = 3.33ALIDD591 pKa = 4.43PKK593 pKa = 11.32SDD595 pKa = 3.26FDD597 pKa = 3.84NLKK600 pKa = 10.76DD601 pKa = 3.16ITIPQEE607 pKa = 4.27SIIKK611 pKa = 10.16LIRR614 pKa = 11.84EE615 pKa = 4.01VGKK618 pKa = 10.56VLNGEE623 pKa = 4.3SLALQYY629 pKa = 10.24PYY631 pKa = 11.42SKK633 pKa = 10.06MDD635 pKa = 3.77DD636 pKa = 3.33RR637 pKa = 11.84GEE639 pKa = 4.19IYY641 pKa = 10.54DD642 pKa = 4.27DD643 pKa = 3.91YY644 pKa = 11.43LVEE647 pKa = 4.25FANVIANTAYY657 pKa = 9.39TKK659 pKa = 11.02AKK661 pKa = 10.17ALEE664 pKa = 4.19ANMMNLISKK673 pKa = 7.38EE674 pKa = 3.88AKK676 pKa = 9.21NLYY679 pKa = 10.45NSFEE683 pKa = 4.16KK684 pKa = 11.04SQDD687 pKa = 3.21PTLASAIEE695 pKa = 4.42GWFEE699 pKa = 4.15DD700 pKa = 4.06LLYY703 pKa = 10.91SDD705 pKa = 5.01GVFDD709 pKa = 4.72IDD711 pKa = 4.14EE712 pKa = 4.63LVDD715 pKa = 4.28SIEE718 pKa = 4.39SNKK721 pKa = 10.04AFYY724 pKa = 10.4YY725 pKa = 10.3RR726 pKa = 11.84QLDD729 pKa = 3.68MEE731 pKa = 4.53KK732 pKa = 10.65ALAIEE737 pKa = 4.15DD738 pKa = 3.78QVNKK742 pKa = 10.41FIFNYY747 pKa = 9.95EE748 pKa = 3.85LTPQEE753 pKa = 4.11FNSINVEE760 pKa = 3.89SAPIMKK766 pKa = 9.71ILSKK770 pKa = 10.53IVGGHH775 pKa = 4.61SSRR778 pKa = 11.84YY779 pKa = 10.18LKK781 pKa = 10.14IQRR784 pKa = 11.84PNN786 pKa = 2.89
MM1 pKa = 7.76KK2 pKa = 9.36ITRR5 pKa = 11.84TYY7 pKa = 11.31KK8 pKa = 10.6NLNSLLKK15 pKa = 10.68SSIVKK20 pKa = 7.38TKK22 pKa = 10.85KK23 pKa = 9.28MFSIRR28 pKa = 11.84DD29 pKa = 3.78SISNSLSIPLRR40 pKa = 11.84QIQYY44 pKa = 10.93LSFGKK49 pKa = 10.05IRR51 pKa = 11.84GLANRR56 pKa = 11.84ALITRR61 pKa = 11.84DD62 pKa = 2.91FMTFVLKK69 pKa = 10.01MKK71 pKa = 9.87KK72 pKa = 9.35NHH74 pKa = 6.37GADD77 pKa = 4.26FTIKK81 pKa = 9.34WLKK84 pKa = 8.89CCYY87 pKa = 9.82VALQKK92 pKa = 11.13SLGKK96 pKa = 10.45DD97 pKa = 3.02NLEE100 pKa = 4.03SLRR103 pKa = 11.84DD104 pKa = 3.83LEE106 pKa = 4.96PNLPLPRR113 pKa = 11.84LINGIPAIISSNDD126 pKa = 2.94RR127 pKa = 11.84KK128 pKa = 10.47LIRR131 pKa = 11.84EE132 pKa = 3.89MDD134 pKa = 3.56SRR136 pKa = 11.84IIIYY140 pKa = 9.45WSSLFSLYY148 pKa = 10.18RR149 pKa = 11.84VLKK152 pKa = 10.38CSYY155 pKa = 9.35KK156 pKa = 10.85LKK158 pKa = 10.29ISSITDD164 pKa = 3.46PFKK167 pKa = 11.33GSINEE172 pKa = 4.11FNDD175 pKa = 3.92IIQGPVFSIFFDD187 pKa = 3.6RR188 pKa = 11.84LNGFPEE194 pKa = 4.29LVKK197 pKa = 10.73KK198 pKa = 11.08SNLAPSKK205 pKa = 10.62VRR207 pKa = 11.84LLRR210 pKa = 11.84SSSSSNNVSWHH221 pKa = 6.44GIITDD226 pKa = 3.07SWNISNSKK234 pKa = 9.86EE235 pKa = 3.69MSSNVSNYY243 pKa = 9.54LASIKK248 pKa = 10.72ALGWNTMWFNSKK260 pKa = 10.03LNEE263 pKa = 4.17MIEE266 pKa = 4.22LGDD269 pKa = 3.86RR270 pKa = 11.84LNQVGSLKK278 pKa = 9.81TKK280 pKa = 10.44KK281 pKa = 10.31SLSGQFGQFSLKK293 pKa = 10.46EE294 pKa = 3.73EE295 pKa = 4.19AAGKK299 pKa = 10.03LRR301 pKa = 11.84IFAIVDD307 pKa = 4.02SITQSLLSPLHH318 pKa = 7.02DD319 pKa = 4.61FMFDD323 pKa = 3.56LLKK326 pKa = 10.64KK327 pKa = 10.03IPNDD331 pKa = 3.47GTFDD335 pKa = 3.24QDD337 pKa = 3.69LSVKK341 pKa = 10.13RR342 pKa = 11.84SQVKK346 pKa = 10.2SLSSGKK352 pKa = 10.26AFSFDD357 pKa = 3.28LSAATDD363 pKa = 3.94RR364 pKa = 11.84LPVDD368 pKa = 3.99LTVKK372 pKa = 10.03ILSKK376 pKa = 10.49IFSDD380 pKa = 4.48EE381 pKa = 4.98FGTSWKK387 pKa = 10.27QLMVNRR393 pKa = 11.84DD394 pKa = 3.51FFFSLTNQKK403 pKa = 10.99DD404 pKa = 3.48YY405 pKa = 11.34GAPEE409 pKa = 3.99SLRR412 pKa = 11.84YY413 pKa = 9.59SVGQPMGALSSWPALALTHH432 pKa = 6.47HH433 pKa = 7.57WILQYY438 pKa = 10.7CSNILGRR445 pKa = 11.84TGWEE449 pKa = 3.65EE450 pKa = 3.73NYY452 pKa = 10.47EE453 pKa = 4.03ILGDD457 pKa = 3.9DD458 pKa = 4.07LVVFDD463 pKa = 5.41SALADD468 pKa = 3.65KK469 pKa = 10.42YY470 pKa = 11.59LEE472 pKa = 3.89IAKK475 pKa = 10.24ILGVEE480 pKa = 3.95INLTKK485 pKa = 10.57SISSHH490 pKa = 5.94DD491 pKa = 3.43RR492 pKa = 11.84PVFEE496 pKa = 4.56FAKK499 pKa = 9.26RR500 pKa = 11.84TCFGNSDD507 pKa = 4.53LSPISIKK514 pKa = 10.42QLLSNDD520 pKa = 3.48QLSEE524 pKa = 3.63RR525 pKa = 11.84TMNVVSFLKK534 pKa = 10.55RR535 pKa = 11.84GLLVSRR541 pKa = 11.84SHH543 pKa = 8.04IGILLSKK550 pKa = 10.18FGSWQILKK558 pKa = 10.26NKK560 pKa = 9.58KK561 pKa = 9.56LSKK564 pKa = 9.81TPLLAILGILNSMNLISHH582 pKa = 6.94RR583 pKa = 11.84WLTEE587 pKa = 3.33ALIDD591 pKa = 4.43PKK593 pKa = 11.32SDD595 pKa = 3.26FDD597 pKa = 3.84NLKK600 pKa = 10.76DD601 pKa = 3.16ITIPQEE607 pKa = 4.27SIIKK611 pKa = 10.16LIRR614 pKa = 11.84EE615 pKa = 4.01VGKK618 pKa = 10.56VLNGEE623 pKa = 4.3SLALQYY629 pKa = 10.24PYY631 pKa = 11.42SKK633 pKa = 10.06MDD635 pKa = 3.77DD636 pKa = 3.33RR637 pKa = 11.84GEE639 pKa = 4.19IYY641 pKa = 10.54DD642 pKa = 4.27DD643 pKa = 3.91YY644 pKa = 11.43LVEE647 pKa = 4.25FANVIANTAYY657 pKa = 9.39TKK659 pKa = 11.02AKK661 pKa = 10.17ALEE664 pKa = 4.19ANMMNLISKK673 pKa = 7.38EE674 pKa = 3.88AKK676 pKa = 9.21NLYY679 pKa = 10.45NSFEE683 pKa = 4.16KK684 pKa = 11.04SQDD687 pKa = 3.21PTLASAIEE695 pKa = 4.42GWFEE699 pKa = 4.15DD700 pKa = 4.06LLYY703 pKa = 10.91SDD705 pKa = 5.01GVFDD709 pKa = 4.72IDD711 pKa = 4.14EE712 pKa = 4.63LVDD715 pKa = 4.28SIEE718 pKa = 4.39SNKK721 pKa = 10.04AFYY724 pKa = 10.4YY725 pKa = 10.3RR726 pKa = 11.84QLDD729 pKa = 3.68MEE731 pKa = 4.53KK732 pKa = 10.65ALAIEE737 pKa = 4.15DD738 pKa = 3.78QVNKK742 pKa = 10.41FIFNYY747 pKa = 9.95EE748 pKa = 3.85LTPQEE753 pKa = 4.11FNSINVEE760 pKa = 3.89SAPIMKK766 pKa = 9.71ILSKK770 pKa = 10.53IVGGHH775 pKa = 4.61SSRR778 pKa = 11.84YY779 pKa = 10.18LKK781 pKa = 10.14IQRR784 pKa = 11.84PNN786 pKa = 2.89
Molecular weight: 89.12 kDa
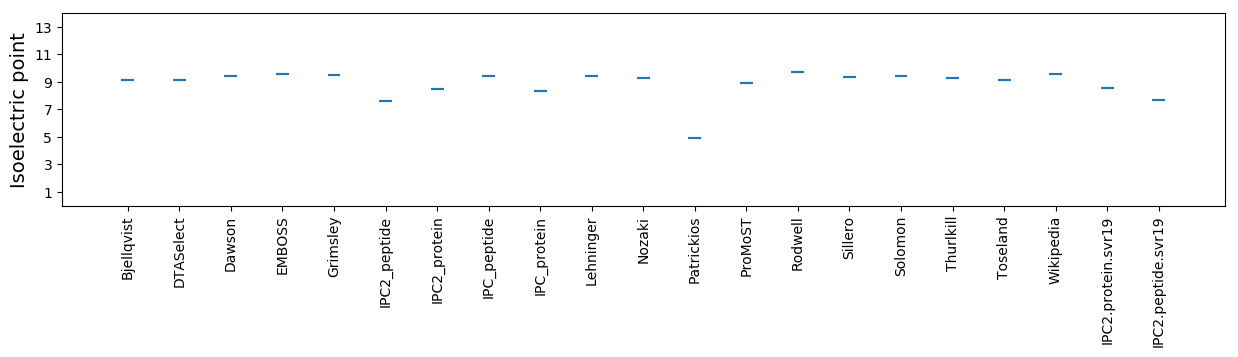
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S4GAV3|A0A0S4GAV3_9VIRU RNA dependent RNA polymerase OS=Botrytis cinerea mitovirus 3 OX=1629666 GN=RdRp PE=4 SV=1
MM1 pKa = 7.76KK2 pKa = 9.36ITRR5 pKa = 11.84TYY7 pKa = 11.31KK8 pKa = 10.6NLNSLLKK15 pKa = 10.68SSIVKK20 pKa = 7.38TKK22 pKa = 10.85KK23 pKa = 9.28MFSIRR28 pKa = 11.84DD29 pKa = 3.78SISNSLSIPLRR40 pKa = 11.84QIQYY44 pKa = 10.93LSFGKK49 pKa = 10.05IRR51 pKa = 11.84GLANRR56 pKa = 11.84ALITRR61 pKa = 11.84DD62 pKa = 2.91FMTFVLKK69 pKa = 10.01MKK71 pKa = 9.87KK72 pKa = 9.35NHH74 pKa = 6.37GADD77 pKa = 4.26FTIKK81 pKa = 9.34WLKK84 pKa = 8.89CCYY87 pKa = 9.82VALQKK92 pKa = 11.13SLGKK96 pKa = 10.45DD97 pKa = 3.02NLEE100 pKa = 4.03SLRR103 pKa = 11.84DD104 pKa = 3.83LEE106 pKa = 4.96PNLPLPRR113 pKa = 11.84LINGIPAIISSNDD126 pKa = 2.94RR127 pKa = 11.84KK128 pKa = 10.47LIRR131 pKa = 11.84EE132 pKa = 3.89MDD134 pKa = 3.56SRR136 pKa = 11.84IIIYY140 pKa = 9.45WSSLFSLYY148 pKa = 10.18RR149 pKa = 11.84VLKK152 pKa = 10.38CSYY155 pKa = 9.35KK156 pKa = 10.85LKK158 pKa = 10.29ISSITDD164 pKa = 3.46PFKK167 pKa = 11.33GSINEE172 pKa = 4.11FNDD175 pKa = 3.92IIQGPVFSIFFDD187 pKa = 3.6RR188 pKa = 11.84LNGFPEE194 pKa = 4.29LVKK197 pKa = 10.73KK198 pKa = 11.08SNLAPSKK205 pKa = 10.62VRR207 pKa = 11.84LLRR210 pKa = 11.84SSSSSNNVSWHH221 pKa = 6.44GIITDD226 pKa = 3.07SWNISNSKK234 pKa = 9.86EE235 pKa = 3.69MSSNVSNYY243 pKa = 9.54LASIKK248 pKa = 10.72ALGWNTMWFNSKK260 pKa = 10.03LNEE263 pKa = 4.17MIEE266 pKa = 4.22LGDD269 pKa = 3.86RR270 pKa = 11.84LNQVGSLKK278 pKa = 9.81TKK280 pKa = 10.44KK281 pKa = 10.31SLSGQFGQFSLKK293 pKa = 10.46EE294 pKa = 3.73EE295 pKa = 4.19AAGKK299 pKa = 10.03LRR301 pKa = 11.84IFAIVDD307 pKa = 4.02SITQSLLSPLHH318 pKa = 7.02DD319 pKa = 4.61FMFDD323 pKa = 3.56LLKK326 pKa = 10.64KK327 pKa = 10.03IPNDD331 pKa = 3.47GTFDD335 pKa = 3.24QDD337 pKa = 3.69LSVKK341 pKa = 10.13RR342 pKa = 11.84SQVKK346 pKa = 10.2SLSSGKK352 pKa = 10.26AFSFDD357 pKa = 3.28LSAATDD363 pKa = 3.94RR364 pKa = 11.84LPVDD368 pKa = 3.99LTVKK372 pKa = 10.03ILSKK376 pKa = 10.49IFSDD380 pKa = 4.48EE381 pKa = 4.98FGTSWKK387 pKa = 10.27QLMVNRR393 pKa = 11.84DD394 pKa = 3.51FFFSLTNQKK403 pKa = 10.99DD404 pKa = 3.48YY405 pKa = 11.34GAPEE409 pKa = 3.99SLRR412 pKa = 11.84YY413 pKa = 9.59SVGQPMGALSSWPALALTHH432 pKa = 6.47HH433 pKa = 7.57WILQYY438 pKa = 10.7CSNILGRR445 pKa = 11.84TGWEE449 pKa = 3.65EE450 pKa = 3.73NYY452 pKa = 10.47EE453 pKa = 4.03ILGDD457 pKa = 3.9DD458 pKa = 4.07LVVFDD463 pKa = 5.41SALADD468 pKa = 3.65KK469 pKa = 10.42YY470 pKa = 11.59LEE472 pKa = 3.89IAKK475 pKa = 10.24ILGVEE480 pKa = 3.95INLTKK485 pKa = 10.57SISSHH490 pKa = 5.94DD491 pKa = 3.43RR492 pKa = 11.84PVFEE496 pKa = 4.56FAKK499 pKa = 9.26RR500 pKa = 11.84TCFGNSDD507 pKa = 4.53LSPISIKK514 pKa = 10.42QLLSNDD520 pKa = 3.48QLSEE524 pKa = 3.63RR525 pKa = 11.84TMNVVSFLKK534 pKa = 10.55RR535 pKa = 11.84GLLVSRR541 pKa = 11.84SHH543 pKa = 8.04IGILLSKK550 pKa = 10.18FGSWQILKK558 pKa = 10.26NKK560 pKa = 9.58KK561 pKa = 9.56LSKK564 pKa = 9.81TPLLAILGILNSMNLISHH582 pKa = 6.94RR583 pKa = 11.84WLTEE587 pKa = 3.33ALIDD591 pKa = 4.43PKK593 pKa = 11.32SDD595 pKa = 3.26FDD597 pKa = 3.84NLKK600 pKa = 10.76DD601 pKa = 3.16ITIPQEE607 pKa = 4.27SIIKK611 pKa = 10.16LIRR614 pKa = 11.84EE615 pKa = 4.01VGKK618 pKa = 10.56VLNGEE623 pKa = 4.3SLALQYY629 pKa = 10.24PYY631 pKa = 11.42SKK633 pKa = 10.06MDD635 pKa = 3.77DD636 pKa = 3.33RR637 pKa = 11.84GEE639 pKa = 4.19IYY641 pKa = 10.54DD642 pKa = 4.27DD643 pKa = 3.91YY644 pKa = 11.43LVEE647 pKa = 4.25FANVIANTAYY657 pKa = 9.39TKK659 pKa = 11.02AKK661 pKa = 10.17ALEE664 pKa = 4.19ANMMNLISKK673 pKa = 7.38EE674 pKa = 3.88AKK676 pKa = 9.21NLYY679 pKa = 10.45NSFEE683 pKa = 4.16KK684 pKa = 11.04SQDD687 pKa = 3.21PTLASAIEE695 pKa = 4.42GWFEE699 pKa = 4.15DD700 pKa = 4.06LLYY703 pKa = 10.91SDD705 pKa = 5.01GVFDD709 pKa = 4.72IDD711 pKa = 4.14EE712 pKa = 4.63LVDD715 pKa = 4.28SIEE718 pKa = 4.39SNKK721 pKa = 10.04AFYY724 pKa = 10.4YY725 pKa = 10.3RR726 pKa = 11.84QLDD729 pKa = 3.68MEE731 pKa = 4.53KK732 pKa = 10.65ALAIEE737 pKa = 4.15DD738 pKa = 3.78QVNKK742 pKa = 10.41FIFNYY747 pKa = 9.95EE748 pKa = 3.85LTPQEE753 pKa = 4.11FNSINVEE760 pKa = 3.89SAPIMKK766 pKa = 9.71ILSKK770 pKa = 10.53IVGGHH775 pKa = 4.61SSRR778 pKa = 11.84YY779 pKa = 10.18LKK781 pKa = 10.14IQRR784 pKa = 11.84PNN786 pKa = 2.89
MM1 pKa = 7.76KK2 pKa = 9.36ITRR5 pKa = 11.84TYY7 pKa = 11.31KK8 pKa = 10.6NLNSLLKK15 pKa = 10.68SSIVKK20 pKa = 7.38TKK22 pKa = 10.85KK23 pKa = 9.28MFSIRR28 pKa = 11.84DD29 pKa = 3.78SISNSLSIPLRR40 pKa = 11.84QIQYY44 pKa = 10.93LSFGKK49 pKa = 10.05IRR51 pKa = 11.84GLANRR56 pKa = 11.84ALITRR61 pKa = 11.84DD62 pKa = 2.91FMTFVLKK69 pKa = 10.01MKK71 pKa = 9.87KK72 pKa = 9.35NHH74 pKa = 6.37GADD77 pKa = 4.26FTIKK81 pKa = 9.34WLKK84 pKa = 8.89CCYY87 pKa = 9.82VALQKK92 pKa = 11.13SLGKK96 pKa = 10.45DD97 pKa = 3.02NLEE100 pKa = 4.03SLRR103 pKa = 11.84DD104 pKa = 3.83LEE106 pKa = 4.96PNLPLPRR113 pKa = 11.84LINGIPAIISSNDD126 pKa = 2.94RR127 pKa = 11.84KK128 pKa = 10.47LIRR131 pKa = 11.84EE132 pKa = 3.89MDD134 pKa = 3.56SRR136 pKa = 11.84IIIYY140 pKa = 9.45WSSLFSLYY148 pKa = 10.18RR149 pKa = 11.84VLKK152 pKa = 10.38CSYY155 pKa = 9.35KK156 pKa = 10.85LKK158 pKa = 10.29ISSITDD164 pKa = 3.46PFKK167 pKa = 11.33GSINEE172 pKa = 4.11FNDD175 pKa = 3.92IIQGPVFSIFFDD187 pKa = 3.6RR188 pKa = 11.84LNGFPEE194 pKa = 4.29LVKK197 pKa = 10.73KK198 pKa = 11.08SNLAPSKK205 pKa = 10.62VRR207 pKa = 11.84LLRR210 pKa = 11.84SSSSSNNVSWHH221 pKa = 6.44GIITDD226 pKa = 3.07SWNISNSKK234 pKa = 9.86EE235 pKa = 3.69MSSNVSNYY243 pKa = 9.54LASIKK248 pKa = 10.72ALGWNTMWFNSKK260 pKa = 10.03LNEE263 pKa = 4.17MIEE266 pKa = 4.22LGDD269 pKa = 3.86RR270 pKa = 11.84LNQVGSLKK278 pKa = 9.81TKK280 pKa = 10.44KK281 pKa = 10.31SLSGQFGQFSLKK293 pKa = 10.46EE294 pKa = 3.73EE295 pKa = 4.19AAGKK299 pKa = 10.03LRR301 pKa = 11.84IFAIVDD307 pKa = 4.02SITQSLLSPLHH318 pKa = 7.02DD319 pKa = 4.61FMFDD323 pKa = 3.56LLKK326 pKa = 10.64KK327 pKa = 10.03IPNDD331 pKa = 3.47GTFDD335 pKa = 3.24QDD337 pKa = 3.69LSVKK341 pKa = 10.13RR342 pKa = 11.84SQVKK346 pKa = 10.2SLSSGKK352 pKa = 10.26AFSFDD357 pKa = 3.28LSAATDD363 pKa = 3.94RR364 pKa = 11.84LPVDD368 pKa = 3.99LTVKK372 pKa = 10.03ILSKK376 pKa = 10.49IFSDD380 pKa = 4.48EE381 pKa = 4.98FGTSWKK387 pKa = 10.27QLMVNRR393 pKa = 11.84DD394 pKa = 3.51FFFSLTNQKK403 pKa = 10.99DD404 pKa = 3.48YY405 pKa = 11.34GAPEE409 pKa = 3.99SLRR412 pKa = 11.84YY413 pKa = 9.59SVGQPMGALSSWPALALTHH432 pKa = 6.47HH433 pKa = 7.57WILQYY438 pKa = 10.7CSNILGRR445 pKa = 11.84TGWEE449 pKa = 3.65EE450 pKa = 3.73NYY452 pKa = 10.47EE453 pKa = 4.03ILGDD457 pKa = 3.9DD458 pKa = 4.07LVVFDD463 pKa = 5.41SALADD468 pKa = 3.65KK469 pKa = 10.42YY470 pKa = 11.59LEE472 pKa = 3.89IAKK475 pKa = 10.24ILGVEE480 pKa = 3.95INLTKK485 pKa = 10.57SISSHH490 pKa = 5.94DD491 pKa = 3.43RR492 pKa = 11.84PVFEE496 pKa = 4.56FAKK499 pKa = 9.26RR500 pKa = 11.84TCFGNSDD507 pKa = 4.53LSPISIKK514 pKa = 10.42QLLSNDD520 pKa = 3.48QLSEE524 pKa = 3.63RR525 pKa = 11.84TMNVVSFLKK534 pKa = 10.55RR535 pKa = 11.84GLLVSRR541 pKa = 11.84SHH543 pKa = 8.04IGILLSKK550 pKa = 10.18FGSWQILKK558 pKa = 10.26NKK560 pKa = 9.58KK561 pKa = 9.56LSKK564 pKa = 9.81TPLLAILGILNSMNLISHH582 pKa = 6.94RR583 pKa = 11.84WLTEE587 pKa = 3.33ALIDD591 pKa = 4.43PKK593 pKa = 11.32SDD595 pKa = 3.26FDD597 pKa = 3.84NLKK600 pKa = 10.76DD601 pKa = 3.16ITIPQEE607 pKa = 4.27SIIKK611 pKa = 10.16LIRR614 pKa = 11.84EE615 pKa = 4.01VGKK618 pKa = 10.56VLNGEE623 pKa = 4.3SLALQYY629 pKa = 10.24PYY631 pKa = 11.42SKK633 pKa = 10.06MDD635 pKa = 3.77DD636 pKa = 3.33RR637 pKa = 11.84GEE639 pKa = 4.19IYY641 pKa = 10.54DD642 pKa = 4.27DD643 pKa = 3.91YY644 pKa = 11.43LVEE647 pKa = 4.25FANVIANTAYY657 pKa = 9.39TKK659 pKa = 11.02AKK661 pKa = 10.17ALEE664 pKa = 4.19ANMMNLISKK673 pKa = 7.38EE674 pKa = 3.88AKK676 pKa = 9.21NLYY679 pKa = 10.45NSFEE683 pKa = 4.16KK684 pKa = 11.04SQDD687 pKa = 3.21PTLASAIEE695 pKa = 4.42GWFEE699 pKa = 4.15DD700 pKa = 4.06LLYY703 pKa = 10.91SDD705 pKa = 5.01GVFDD709 pKa = 4.72IDD711 pKa = 4.14EE712 pKa = 4.63LVDD715 pKa = 4.28SIEE718 pKa = 4.39SNKK721 pKa = 10.04AFYY724 pKa = 10.4YY725 pKa = 10.3RR726 pKa = 11.84QLDD729 pKa = 3.68MEE731 pKa = 4.53KK732 pKa = 10.65ALAIEE737 pKa = 4.15DD738 pKa = 3.78QVNKK742 pKa = 10.41FIFNYY747 pKa = 9.95EE748 pKa = 3.85LTPQEE753 pKa = 4.11FNSINVEE760 pKa = 3.89SAPIMKK766 pKa = 9.71ILSKK770 pKa = 10.53IVGGHH775 pKa = 4.61SSRR778 pKa = 11.84YY779 pKa = 10.18LKK781 pKa = 10.14IQRR784 pKa = 11.84PNN786 pKa = 2.89
Molecular weight: 89.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
786 |
786 |
786 |
786.0 |
89.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.835 ± 0.0 | 0.636 ± 0.0 |
5.98 ± 0.0 | 4.707 ± 0.0 |
5.216 ± 0.0 | 4.707 ± 0.0 |
1.145 ± 0.0 | 8.651 ± 0.0 |
8.397 ± 0.0 | 12.468 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.29 ± 0.0 | 6.234 ± 0.0 |
3.181 ± 0.0 | 3.053 ± 0.0 |
4.198 ± 0.0 | 11.959 ± 0.0 |
3.562 ± 0.0 | 4.198 ± 0.0 |
1.654 ± 0.0 | 2.926 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
