
Lettuce necrotic yellows virus (isolate 318) (LNYV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Lettuce necrotic yellows cytorhabdovirus; Lettuce necrotic yellows virus
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
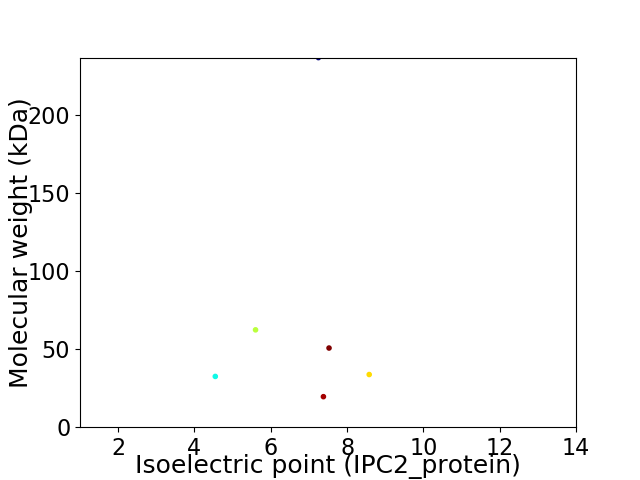
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9E7N8|MVP_LNYV3 Probable movement protein 4b OS=Lettuce necrotic yellows virus (isolate 318) OX=928304 GN=4b PE=2 SV=2
MM1 pKa = 8.03DD2 pKa = 4.95SEE4 pKa = 4.96SLDD7 pKa = 3.54FSSADD12 pKa = 3.57TVILRR17 pKa = 11.84SPNAGTNPDD26 pKa = 3.46GHH28 pKa = 7.43PDD30 pKa = 3.45TVEE33 pKa = 4.52CPDD36 pKa = 4.38FDD38 pKa = 4.08TDD40 pKa = 4.23IPKK43 pKa = 10.12TSDD46 pKa = 3.85DD47 pKa = 3.82SSKK50 pKa = 10.37MDD52 pKa = 3.48NKK54 pKa = 10.64GSSSSSKK61 pKa = 10.44AVKK64 pKa = 10.33DD65 pKa = 3.89LLEE68 pKa = 4.52LAAKK72 pKa = 9.74SQGIVVTDD80 pKa = 3.14VMQNTAIALHH90 pKa = 6.63HH91 pKa = 6.33NLGLDD96 pKa = 3.41ASSLDD101 pKa = 3.37WFVAGITFANNSMIMEE117 pKa = 4.37KK118 pKa = 9.48MVSAIKK124 pKa = 10.01EE125 pKa = 3.92LQIEE129 pKa = 4.18VRR131 pKa = 11.84NIQVASSGIKK141 pKa = 8.6GTSEE145 pKa = 3.81EE146 pKa = 4.78LVSKK150 pKa = 10.29MKK152 pKa = 10.72ANKK155 pKa = 9.61NDD157 pKa = 3.28IVKK160 pKa = 10.12EE161 pKa = 3.9LVKK164 pKa = 10.31TRR166 pKa = 11.84DD167 pKa = 3.57SVLSAMGGILSAPEE181 pKa = 4.26IEE183 pKa = 4.29QQPVKK188 pKa = 9.81TVTIGASQGRR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 9.09STVVPPIEE208 pKa = 4.41INPEE212 pKa = 3.84LEE214 pKa = 4.19SPVLSKK220 pKa = 9.69TVSTATPEE228 pKa = 3.5EE229 pKa = 4.59RR230 pKa = 11.84IRR232 pKa = 11.84HH233 pKa = 4.98EE234 pKa = 4.18KK235 pKa = 10.4EE236 pKa = 3.59KK237 pKa = 11.09LLADD241 pKa = 4.74LDD243 pKa = 4.06WEE245 pKa = 4.25IGEE248 pKa = 4.24IAQYY252 pKa = 10.35TPLIVDD258 pKa = 4.25FLVPDD263 pKa = 5.83DD264 pKa = 4.4ILAMAADD271 pKa = 4.35GLTPEE276 pKa = 4.88LKK278 pKa = 10.5EE279 pKa = 4.29KK280 pKa = 10.02IQNEE284 pKa = 3.99IIEE287 pKa = 4.07NHH289 pKa = 5.78IALMALEE296 pKa = 5.18EE297 pKa = 4.29YY298 pKa = 10.97SSS300 pKa = 3.38
MM1 pKa = 8.03DD2 pKa = 4.95SEE4 pKa = 4.96SLDD7 pKa = 3.54FSSADD12 pKa = 3.57TVILRR17 pKa = 11.84SPNAGTNPDD26 pKa = 3.46GHH28 pKa = 7.43PDD30 pKa = 3.45TVEE33 pKa = 4.52CPDD36 pKa = 4.38FDD38 pKa = 4.08TDD40 pKa = 4.23IPKK43 pKa = 10.12TSDD46 pKa = 3.85DD47 pKa = 3.82SSKK50 pKa = 10.37MDD52 pKa = 3.48NKK54 pKa = 10.64GSSSSSKK61 pKa = 10.44AVKK64 pKa = 10.33DD65 pKa = 3.89LLEE68 pKa = 4.52LAAKK72 pKa = 9.74SQGIVVTDD80 pKa = 3.14VMQNTAIALHH90 pKa = 6.63HH91 pKa = 6.33NLGLDD96 pKa = 3.41ASSLDD101 pKa = 3.37WFVAGITFANNSMIMEE117 pKa = 4.37KK118 pKa = 9.48MVSAIKK124 pKa = 10.01EE125 pKa = 3.92LQIEE129 pKa = 4.18VRR131 pKa = 11.84NIQVASSGIKK141 pKa = 8.6GTSEE145 pKa = 3.81EE146 pKa = 4.78LVSKK150 pKa = 10.29MKK152 pKa = 10.72ANKK155 pKa = 9.61NDD157 pKa = 3.28IVKK160 pKa = 10.12EE161 pKa = 3.9LVKK164 pKa = 10.31TRR166 pKa = 11.84DD167 pKa = 3.57SVLSAMGGILSAPEE181 pKa = 4.26IEE183 pKa = 4.29QQPVKK188 pKa = 9.81TVTIGASQGRR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 9.09STVVPPIEE208 pKa = 4.41INPEE212 pKa = 3.84LEE214 pKa = 4.19SPVLSKK220 pKa = 9.69TVSTATPEE228 pKa = 3.5EE229 pKa = 4.59RR230 pKa = 11.84IRR232 pKa = 11.84HH233 pKa = 4.98EE234 pKa = 4.18KK235 pKa = 10.4EE236 pKa = 3.59KK237 pKa = 11.09LLADD241 pKa = 4.74LDD243 pKa = 4.06WEE245 pKa = 4.25IGEE248 pKa = 4.24IAQYY252 pKa = 10.35TPLIVDD258 pKa = 4.25FLVPDD263 pKa = 5.83DD264 pKa = 4.4ILAMAADD271 pKa = 4.35GLTPEE276 pKa = 4.88LKK278 pKa = 10.5EE279 pKa = 4.29KK280 pKa = 10.02IQNEE284 pKa = 3.99IIEE287 pKa = 4.07NHH289 pKa = 5.78IALMALEE296 pKa = 5.18EE297 pKa = 4.29YY298 pKa = 10.97SSS300 pKa = 3.38
Molecular weight: 32.5 kDa
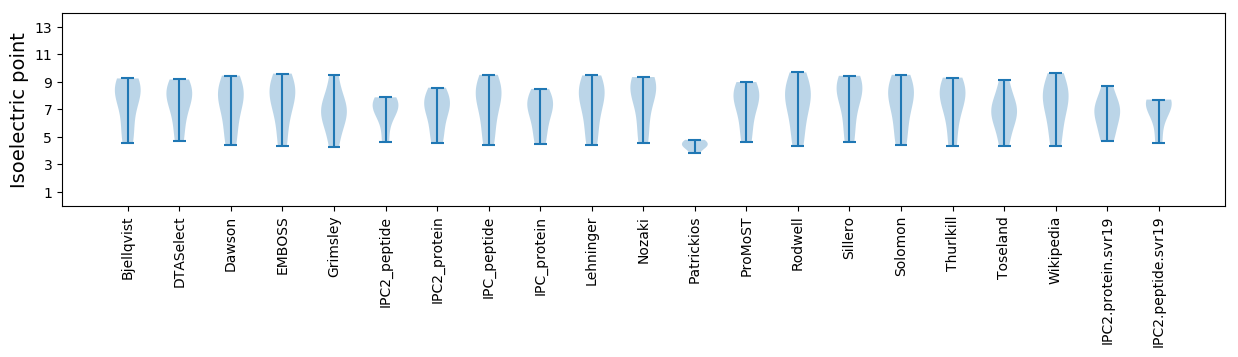
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9E7N9|MATRX_LNYV3 Matrix protein OS=Lettuce necrotic yellows virus (isolate 318) OX=928304 GN=M PE=2 SV=1
MM1 pKa = 7.73LDD3 pKa = 3.32VNSVRR8 pKa = 11.84KK9 pKa = 9.19HH10 pKa = 4.16VLKK13 pKa = 10.18TGSLTSAVGTGTIYY27 pKa = 10.21QGTYY31 pKa = 9.13NRR33 pKa = 11.84YY34 pKa = 9.34AKK36 pKa = 10.37KK37 pKa = 10.31KK38 pKa = 8.14EE39 pKa = 3.71LNIIVTSSGSNNVIMRR55 pKa = 11.84QVPLFDD61 pKa = 5.88KK62 pKa = 10.89EE63 pKa = 5.17DD64 pKa = 3.96LDD66 pKa = 4.72AMKK69 pKa = 10.79SDD71 pKa = 3.69TTSNKK76 pKa = 9.37YY77 pKa = 10.06LHH79 pKa = 6.89IGCITVSIEE88 pKa = 3.81PLLHH92 pKa = 4.94QRR94 pKa = 11.84YY95 pKa = 7.25MKK97 pKa = 10.92NFGKK101 pKa = 9.12TIAGNCAIIDD111 pKa = 3.75STFRR115 pKa = 11.84KK116 pKa = 9.46VDD118 pKa = 3.18QSIISLHH125 pKa = 6.57KK126 pKa = 9.56YY127 pKa = 9.56DD128 pKa = 4.72LSRR131 pKa = 11.84GRR133 pKa = 11.84ADD135 pKa = 3.28YY136 pKa = 11.03VSYY139 pKa = 10.7PNHH142 pKa = 6.6CLSLTDD148 pKa = 3.86PMIQKK153 pKa = 10.16RR154 pKa = 11.84LSVLLGIKK162 pKa = 10.36GIDD165 pKa = 3.48VEE167 pKa = 4.59PGVEE171 pKa = 4.36LFSICIGYY179 pKa = 9.33IVSSVNTLHH188 pKa = 6.97PVSQLGIQGVAINGTEE204 pKa = 3.98SADD207 pKa = 3.38IDD209 pKa = 3.77EE210 pKa = 5.56LGAEE214 pKa = 5.33DD215 pKa = 5.11IDD217 pKa = 4.53QLSLSYY223 pKa = 10.82NDD225 pKa = 3.99SKK227 pKa = 11.08IISLPSDD234 pKa = 2.96EE235 pKa = 5.47DD236 pKa = 3.0IYY238 pKa = 11.47YY239 pKa = 10.34RR240 pKa = 11.84SKK242 pKa = 11.35GSLFSKK248 pKa = 10.43GRR250 pKa = 11.84TIKK253 pKa = 10.32RR254 pKa = 11.84RR255 pKa = 11.84TMRR258 pKa = 11.84TRR260 pKa = 11.84VPDD263 pKa = 3.7PEE265 pKa = 4.27EE266 pKa = 4.59PIKK269 pKa = 10.09LTKK272 pKa = 10.14SQSSRR277 pKa = 11.84IEE279 pKa = 3.7HH280 pKa = 5.65GKK282 pKa = 10.06VMRR285 pKa = 11.84LLKK288 pKa = 10.51NKK290 pKa = 9.59QIRR293 pKa = 11.84EE294 pKa = 4.03KK295 pKa = 10.46IEE297 pKa = 3.92RR298 pKa = 11.84GMIAA302 pKa = 3.92
MM1 pKa = 7.73LDD3 pKa = 3.32VNSVRR8 pKa = 11.84KK9 pKa = 9.19HH10 pKa = 4.16VLKK13 pKa = 10.18TGSLTSAVGTGTIYY27 pKa = 10.21QGTYY31 pKa = 9.13NRR33 pKa = 11.84YY34 pKa = 9.34AKK36 pKa = 10.37KK37 pKa = 10.31KK38 pKa = 8.14EE39 pKa = 3.71LNIIVTSSGSNNVIMRR55 pKa = 11.84QVPLFDD61 pKa = 5.88KK62 pKa = 10.89EE63 pKa = 5.17DD64 pKa = 3.96LDD66 pKa = 4.72AMKK69 pKa = 10.79SDD71 pKa = 3.69TTSNKK76 pKa = 9.37YY77 pKa = 10.06LHH79 pKa = 6.89IGCITVSIEE88 pKa = 3.81PLLHH92 pKa = 4.94QRR94 pKa = 11.84YY95 pKa = 7.25MKK97 pKa = 10.92NFGKK101 pKa = 9.12TIAGNCAIIDD111 pKa = 3.75STFRR115 pKa = 11.84KK116 pKa = 9.46VDD118 pKa = 3.18QSIISLHH125 pKa = 6.57KK126 pKa = 9.56YY127 pKa = 9.56DD128 pKa = 4.72LSRR131 pKa = 11.84GRR133 pKa = 11.84ADD135 pKa = 3.28YY136 pKa = 11.03VSYY139 pKa = 10.7PNHH142 pKa = 6.6CLSLTDD148 pKa = 3.86PMIQKK153 pKa = 10.16RR154 pKa = 11.84LSVLLGIKK162 pKa = 10.36GIDD165 pKa = 3.48VEE167 pKa = 4.59PGVEE171 pKa = 4.36LFSICIGYY179 pKa = 9.33IVSSVNTLHH188 pKa = 6.97PVSQLGIQGVAINGTEE204 pKa = 3.98SADD207 pKa = 3.38IDD209 pKa = 3.77EE210 pKa = 5.56LGAEE214 pKa = 5.33DD215 pKa = 5.11IDD217 pKa = 4.53QLSLSYY223 pKa = 10.82NDD225 pKa = 3.99SKK227 pKa = 11.08IISLPSDD234 pKa = 2.96EE235 pKa = 5.47DD236 pKa = 3.0IYY238 pKa = 11.47YY239 pKa = 10.34RR240 pKa = 11.84SKK242 pKa = 11.35GSLFSKK248 pKa = 10.43GRR250 pKa = 11.84TIKK253 pKa = 10.32RR254 pKa = 11.84RR255 pKa = 11.84TMRR258 pKa = 11.84TRR260 pKa = 11.84VPDD263 pKa = 3.7PEE265 pKa = 4.27EE266 pKa = 4.59PIKK269 pKa = 10.09LTKK272 pKa = 10.14SQSSRR277 pKa = 11.84IEE279 pKa = 3.7HH280 pKa = 5.65GKK282 pKa = 10.06VMRR285 pKa = 11.84LLKK288 pKa = 10.51NKK290 pKa = 9.59QIRR293 pKa = 11.84EE294 pKa = 4.03KK295 pKa = 10.46IEE297 pKa = 3.92RR298 pKa = 11.84GMIAA302 pKa = 3.92
Molecular weight: 33.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3857 |
177 |
2068 |
642.8 |
72.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.6 ± 1.604 | 1.815 ± 0.362 |
6.222 ± 0.363 | 5.963 ± 0.375 |
3.189 ± 0.447 | 5.626 ± 0.309 |
2.126 ± 0.211 | 6.611 ± 0.664 |
6.404 ± 0.264 | 9.437 ± 0.493 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.267 ± 0.393 | 3.837 ± 0.134 |
4.511 ± 0.142 | 2.904 ± 0.434 |
5.548 ± 0.477 | 8.608 ± 0.616 |
6.663 ± 0.52 | 6.482 ± 0.303 |
1.685 ± 0.338 | 3.5 ± 0.426 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
