
Beihai tombus-like virus 13
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.35
Get precalculated fractions of proteins
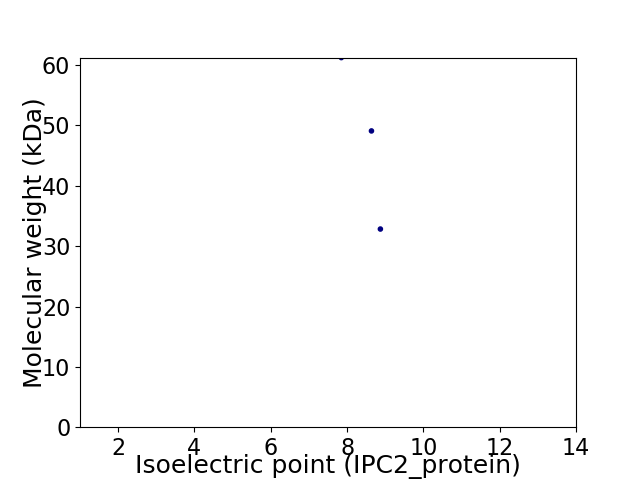
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFV4|A0A1L3KFV4_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 13 OX=1922716 PE=4 SV=1
MM1 pKa = 7.7VPICRR6 pKa = 11.84GVQHH10 pKa = 7.08LLEE13 pKa = 4.31NHH15 pKa = 6.4PQDD18 pKa = 4.32EE19 pKa = 4.49NASPDD24 pKa = 3.69LLLDD28 pKa = 3.42TRR30 pKa = 11.84NLGFHH35 pKa = 6.09RR36 pKa = 11.84QSYY39 pKa = 9.64HH40 pKa = 5.79YY41 pKa = 10.58KK42 pKa = 10.55LFNLPPSMEE51 pKa = 4.28CMLEE55 pKa = 3.83KK56 pKa = 9.83QTFYY60 pKa = 11.59NNCYY64 pKa = 9.13CNEE67 pKa = 3.98YY68 pKa = 9.9NAMIGRR74 pKa = 11.84HH75 pKa = 5.26FTPDD79 pKa = 3.48LPEE82 pKa = 4.7KK83 pKa = 10.76SSIPALTEE91 pKa = 3.68LFEE94 pKa = 4.56KK95 pKa = 10.8EE96 pKa = 3.9NIKK99 pKa = 11.01YY100 pKa = 9.43CDD102 pKa = 3.35SMKK105 pKa = 10.75EE106 pKa = 4.03LLFSTGGYY114 pKa = 9.56NPPSYY119 pKa = 11.16ASVISNTKK127 pKa = 8.28GAKK130 pKa = 8.64KK131 pKa = 9.99KK132 pKa = 10.16RR133 pKa = 11.84YY134 pKa = 8.43QRR136 pKa = 11.84AYY138 pKa = 11.02DD139 pKa = 3.84NILNRR144 pKa = 11.84RR145 pKa = 11.84ILFDD149 pKa = 3.49NRR151 pKa = 11.84HH152 pKa = 5.05SRR154 pKa = 11.84VNFFVKK160 pKa = 10.51LEE162 pKa = 3.8KK163 pKa = 10.21WPISKK168 pKa = 10.03VEE170 pKa = 4.02KK171 pKa = 9.8EE172 pKa = 4.0KK173 pKa = 10.71PPRR176 pKa = 11.84GIQFRR181 pKa = 11.84SYY183 pKa = 11.02EE184 pKa = 3.89YY185 pKa = 11.32LLALKK190 pKa = 10.4RR191 pKa = 11.84LITPMVDD198 pKa = 3.04LTKK201 pKa = 10.15TNKK204 pKa = 8.87TFEE207 pKa = 4.65HH208 pKa = 6.36NPPFNPQCVFTKK220 pKa = 10.84NNTPQVIAANFLEE233 pKa = 4.58AWQKK237 pKa = 10.99YY238 pKa = 5.62KK239 pKa = 10.8TPTAVCLDD247 pKa = 3.82HH248 pKa = 7.36SCFDD252 pKa = 3.24GHH254 pKa = 6.48YY255 pKa = 9.36SHH257 pKa = 7.22EE258 pKa = 4.13LLTYY262 pKa = 10.27EE263 pKa = 3.88MQRR266 pKa = 11.84YY267 pKa = 7.54QRR269 pKa = 11.84ISGCKK274 pKa = 9.26NNSLLMRR281 pKa = 11.84LLKK284 pKa = 10.54RR285 pKa = 11.84QLTNTGYY292 pKa = 9.91SAGGIKK298 pKa = 10.57YY299 pKa = 8.98KK300 pKa = 11.04VKK302 pKa = 10.47GKK304 pKa = 9.69RR305 pKa = 11.84CSGEE309 pKa = 3.71FTTSHH314 pKa = 6.49GNGEE318 pKa = 4.39TNYY321 pKa = 11.24LMIRR325 pKa = 11.84SILLFLGIMEE335 pKa = 4.56FNIFVNGDD343 pKa = 3.5DD344 pKa = 5.02SVIICNFEE352 pKa = 3.95DD353 pKa = 3.17VDD355 pKa = 4.94RR356 pKa = 11.84IVKK359 pKa = 10.03NLEE362 pKa = 3.56LFRR365 pKa = 11.84NFNMSTEE372 pKa = 4.25VEE374 pKa = 4.56HH375 pKa = 6.62IATVFEE381 pKa = 5.27EE382 pKa = 4.07ITFCQTSPVLVNGTYY397 pKa = 11.06KK398 pKa = 9.82MVRR401 pKa = 11.84KK402 pKa = 9.41PMRR405 pKa = 11.84VLSRR409 pKa = 11.84LPYY412 pKa = 10.63SSTNWASVLDD422 pKa = 3.89RR423 pKa = 11.84FMVSVGLCEE432 pKa = 3.99LSINIGVPILQEE444 pKa = 3.6LAIWLIRR451 pKa = 11.84KK452 pKa = 8.84GGSDD456 pKa = 4.33RR457 pKa = 11.84PLTTHH462 pKa = 6.27KK463 pKa = 9.32TDD465 pKa = 3.64HH466 pKa = 6.73YY467 pKa = 11.01DD468 pKa = 3.17AQAVLKK474 pKa = 10.43IEE476 pKa = 5.41NIDD479 pKa = 3.31QSTRR483 pKa = 11.84EE484 pKa = 4.15SFNLAFGIDD493 pKa = 3.43PHH495 pKa = 6.02EE496 pKa = 4.01QLRR499 pKa = 11.84LEE501 pKa = 4.38KK502 pKa = 10.53YY503 pKa = 10.69LRR505 pKa = 11.84DD506 pKa = 4.62DD507 pKa = 4.02YY508 pKa = 11.37CDD510 pKa = 4.39HH511 pKa = 7.88PDD513 pKa = 3.96QIQTLLTKK521 pKa = 10.76LNYY524 pKa = 10.08KK525 pKa = 10.45LYY527 pKa = 10.94
MM1 pKa = 7.7VPICRR6 pKa = 11.84GVQHH10 pKa = 7.08LLEE13 pKa = 4.31NHH15 pKa = 6.4PQDD18 pKa = 4.32EE19 pKa = 4.49NASPDD24 pKa = 3.69LLLDD28 pKa = 3.42TRR30 pKa = 11.84NLGFHH35 pKa = 6.09RR36 pKa = 11.84QSYY39 pKa = 9.64HH40 pKa = 5.79YY41 pKa = 10.58KK42 pKa = 10.55LFNLPPSMEE51 pKa = 4.28CMLEE55 pKa = 3.83KK56 pKa = 9.83QTFYY60 pKa = 11.59NNCYY64 pKa = 9.13CNEE67 pKa = 3.98YY68 pKa = 9.9NAMIGRR74 pKa = 11.84HH75 pKa = 5.26FTPDD79 pKa = 3.48LPEE82 pKa = 4.7KK83 pKa = 10.76SSIPALTEE91 pKa = 3.68LFEE94 pKa = 4.56KK95 pKa = 10.8EE96 pKa = 3.9NIKK99 pKa = 11.01YY100 pKa = 9.43CDD102 pKa = 3.35SMKK105 pKa = 10.75EE106 pKa = 4.03LLFSTGGYY114 pKa = 9.56NPPSYY119 pKa = 11.16ASVISNTKK127 pKa = 8.28GAKK130 pKa = 8.64KK131 pKa = 9.99KK132 pKa = 10.16RR133 pKa = 11.84YY134 pKa = 8.43QRR136 pKa = 11.84AYY138 pKa = 11.02DD139 pKa = 3.84NILNRR144 pKa = 11.84RR145 pKa = 11.84ILFDD149 pKa = 3.49NRR151 pKa = 11.84HH152 pKa = 5.05SRR154 pKa = 11.84VNFFVKK160 pKa = 10.51LEE162 pKa = 3.8KK163 pKa = 10.21WPISKK168 pKa = 10.03VEE170 pKa = 4.02KK171 pKa = 9.8EE172 pKa = 4.0KK173 pKa = 10.71PPRR176 pKa = 11.84GIQFRR181 pKa = 11.84SYY183 pKa = 11.02EE184 pKa = 3.89YY185 pKa = 11.32LLALKK190 pKa = 10.4RR191 pKa = 11.84LITPMVDD198 pKa = 3.04LTKK201 pKa = 10.15TNKK204 pKa = 8.87TFEE207 pKa = 4.65HH208 pKa = 6.36NPPFNPQCVFTKK220 pKa = 10.84NNTPQVIAANFLEE233 pKa = 4.58AWQKK237 pKa = 10.99YY238 pKa = 5.62KK239 pKa = 10.8TPTAVCLDD247 pKa = 3.82HH248 pKa = 7.36SCFDD252 pKa = 3.24GHH254 pKa = 6.48YY255 pKa = 9.36SHH257 pKa = 7.22EE258 pKa = 4.13LLTYY262 pKa = 10.27EE263 pKa = 3.88MQRR266 pKa = 11.84YY267 pKa = 7.54QRR269 pKa = 11.84ISGCKK274 pKa = 9.26NNSLLMRR281 pKa = 11.84LLKK284 pKa = 10.54RR285 pKa = 11.84QLTNTGYY292 pKa = 9.91SAGGIKK298 pKa = 10.57YY299 pKa = 8.98KK300 pKa = 11.04VKK302 pKa = 10.47GKK304 pKa = 9.69RR305 pKa = 11.84CSGEE309 pKa = 3.71FTTSHH314 pKa = 6.49GNGEE318 pKa = 4.39TNYY321 pKa = 11.24LMIRR325 pKa = 11.84SILLFLGIMEE335 pKa = 4.56FNIFVNGDD343 pKa = 3.5DD344 pKa = 5.02SVIICNFEE352 pKa = 3.95DD353 pKa = 3.17VDD355 pKa = 4.94RR356 pKa = 11.84IVKK359 pKa = 10.03NLEE362 pKa = 3.56LFRR365 pKa = 11.84NFNMSTEE372 pKa = 4.25VEE374 pKa = 4.56HH375 pKa = 6.62IATVFEE381 pKa = 5.27EE382 pKa = 4.07ITFCQTSPVLVNGTYY397 pKa = 11.06KK398 pKa = 9.82MVRR401 pKa = 11.84KK402 pKa = 9.41PMRR405 pKa = 11.84VLSRR409 pKa = 11.84LPYY412 pKa = 10.63SSTNWASVLDD422 pKa = 3.89RR423 pKa = 11.84FMVSVGLCEE432 pKa = 3.99LSINIGVPILQEE444 pKa = 3.6LAIWLIRR451 pKa = 11.84KK452 pKa = 8.84GGSDD456 pKa = 4.33RR457 pKa = 11.84PLTTHH462 pKa = 6.27KK463 pKa = 9.32TDD465 pKa = 3.64HH466 pKa = 6.73YY467 pKa = 11.01DD468 pKa = 3.17AQAVLKK474 pKa = 10.43IEE476 pKa = 5.41NIDD479 pKa = 3.31QSTRR483 pKa = 11.84EE484 pKa = 4.15SFNLAFGIDD493 pKa = 3.43PHH495 pKa = 6.02EE496 pKa = 4.01QLRR499 pKa = 11.84LEE501 pKa = 4.38KK502 pKa = 10.53YY503 pKa = 10.69LRR505 pKa = 11.84DD506 pKa = 4.62DD507 pKa = 4.02YY508 pKa = 11.37CDD510 pKa = 4.39HH511 pKa = 7.88PDD513 pKa = 3.96QIQTLLTKK521 pKa = 10.76LNYY524 pKa = 10.08KK525 pKa = 10.45LYY527 pKa = 10.94
Molecular weight: 61.25 kDa
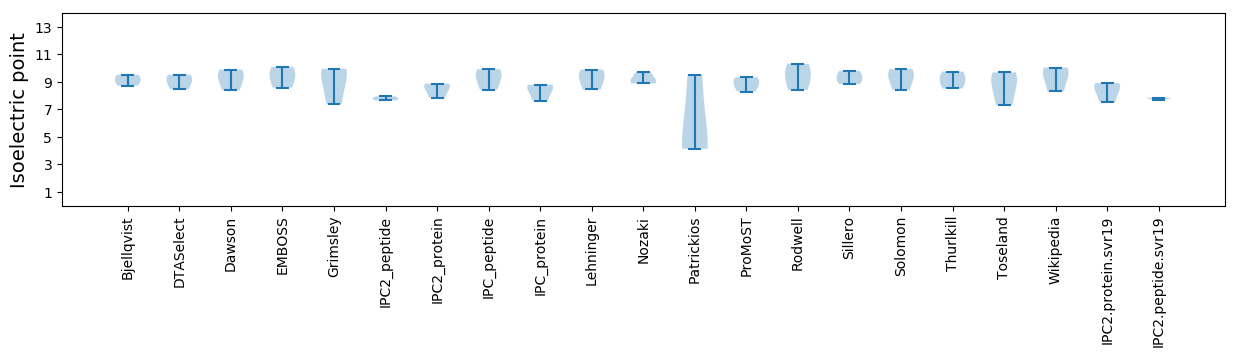
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFV4|A0A1L3KFV4_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 13 OX=1922716 PE=4 SV=1
MM1 pKa = 7.51EE2 pKa = 4.05MAFYY6 pKa = 10.19QIRR9 pKa = 11.84EE10 pKa = 4.18MPPLLARR17 pKa = 11.84IEE19 pKa = 4.12EE20 pKa = 4.34RR21 pKa = 11.84PQRR24 pKa = 11.84RR25 pKa = 11.84AEE27 pKa = 3.98DD28 pKa = 3.81RR29 pKa = 11.84PVLSRR34 pKa = 11.84MFYY37 pKa = 10.71KK38 pKa = 10.4LRR40 pKa = 11.84KK41 pKa = 8.73IKK43 pKa = 10.79NHH45 pKa = 5.19MKK47 pKa = 10.0KK48 pKa = 10.56APAEE52 pKa = 3.96EE53 pKa = 4.64HH54 pKa = 6.66NILPVLQEE62 pKa = 4.2TITPVALHH70 pKa = 6.06NAFSLLHH77 pKa = 6.47DD78 pKa = 5.11AGDD81 pKa = 3.62IVNDD85 pKa = 3.66EE86 pKa = 4.43QIDD89 pKa = 3.83IVLSEE94 pKa = 4.34TPKK97 pKa = 10.44KK98 pKa = 10.73VKK100 pKa = 10.44DD101 pKa = 3.94VIKK104 pKa = 10.37KK105 pKa = 9.58VPKK108 pKa = 9.37AQLNARR114 pKa = 11.84LATVDD119 pKa = 4.04EE120 pKa = 4.63NGLLQTSANFVEE132 pKa = 4.57HH133 pKa = 7.25DD134 pKa = 3.64VAIEE138 pKa = 3.85DD139 pKa = 3.96RR140 pKa = 11.84FEE142 pKa = 3.96MSTDD146 pKa = 3.36RR147 pKa = 11.84EE148 pKa = 4.2RR149 pKa = 11.84IRR151 pKa = 11.84PAKK154 pKa = 9.71RR155 pKa = 11.84IKK157 pKa = 10.83GEE159 pKa = 3.65FRR161 pKa = 11.84KK162 pKa = 10.22AVAAVVKK169 pKa = 10.59SDD171 pKa = 2.91MYY173 pKa = 11.3YY174 pKa = 10.62YY175 pKa = 10.28MKK177 pKa = 10.13IKK179 pKa = 10.76YY180 pKa = 9.24FMKK183 pKa = 10.04EE184 pKa = 3.69RR185 pKa = 11.84TPGMVQQIINDD196 pKa = 3.4CRR198 pKa = 11.84VYY200 pKa = 9.64LTKK203 pKa = 10.59SGSLMNTKK211 pKa = 10.05EE212 pKa = 4.16DD213 pKa = 3.97YY214 pKa = 10.78EE215 pKa = 4.45LVSSLVLAIWLPSRR229 pKa = 11.84EE230 pKa = 3.97EE231 pKa = 3.77LRR233 pKa = 11.84FRR235 pKa = 11.84SAMKK239 pKa = 10.78NNVVLDD245 pKa = 4.5GIEE248 pKa = 5.64AINQFNKK255 pKa = 10.44GNLGNRR261 pKa = 11.84FSIPIPSVSRR271 pKa = 11.84PSSLTKK277 pKa = 10.3QVTINPKK284 pKa = 8.47QQ285 pKa = 3.09
MM1 pKa = 7.51EE2 pKa = 4.05MAFYY6 pKa = 10.19QIRR9 pKa = 11.84EE10 pKa = 4.18MPPLLARR17 pKa = 11.84IEE19 pKa = 4.12EE20 pKa = 4.34RR21 pKa = 11.84PQRR24 pKa = 11.84RR25 pKa = 11.84AEE27 pKa = 3.98DD28 pKa = 3.81RR29 pKa = 11.84PVLSRR34 pKa = 11.84MFYY37 pKa = 10.71KK38 pKa = 10.4LRR40 pKa = 11.84KK41 pKa = 8.73IKK43 pKa = 10.79NHH45 pKa = 5.19MKK47 pKa = 10.0KK48 pKa = 10.56APAEE52 pKa = 3.96EE53 pKa = 4.64HH54 pKa = 6.66NILPVLQEE62 pKa = 4.2TITPVALHH70 pKa = 6.06NAFSLLHH77 pKa = 6.47DD78 pKa = 5.11AGDD81 pKa = 3.62IVNDD85 pKa = 3.66EE86 pKa = 4.43QIDD89 pKa = 3.83IVLSEE94 pKa = 4.34TPKK97 pKa = 10.44KK98 pKa = 10.73VKK100 pKa = 10.44DD101 pKa = 3.94VIKK104 pKa = 10.37KK105 pKa = 9.58VPKK108 pKa = 9.37AQLNARR114 pKa = 11.84LATVDD119 pKa = 4.04EE120 pKa = 4.63NGLLQTSANFVEE132 pKa = 4.57HH133 pKa = 7.25DD134 pKa = 3.64VAIEE138 pKa = 3.85DD139 pKa = 3.96RR140 pKa = 11.84FEE142 pKa = 3.96MSTDD146 pKa = 3.36RR147 pKa = 11.84EE148 pKa = 4.2RR149 pKa = 11.84IRR151 pKa = 11.84PAKK154 pKa = 9.71RR155 pKa = 11.84IKK157 pKa = 10.83GEE159 pKa = 3.65FRR161 pKa = 11.84KK162 pKa = 10.22AVAAVVKK169 pKa = 10.59SDD171 pKa = 2.91MYY173 pKa = 11.3YY174 pKa = 10.62YY175 pKa = 10.28MKK177 pKa = 10.13IKK179 pKa = 10.76YY180 pKa = 9.24FMKK183 pKa = 10.04EE184 pKa = 3.69RR185 pKa = 11.84TPGMVQQIINDD196 pKa = 3.4CRR198 pKa = 11.84VYY200 pKa = 9.64LTKK203 pKa = 10.59SGSLMNTKK211 pKa = 10.05EE212 pKa = 4.16DD213 pKa = 3.97YY214 pKa = 10.78EE215 pKa = 4.45LVSSLVLAIWLPSRR229 pKa = 11.84EE230 pKa = 3.97EE231 pKa = 3.77LRR233 pKa = 11.84FRR235 pKa = 11.84SAMKK239 pKa = 10.78NNVVLDD245 pKa = 4.5GIEE248 pKa = 5.64AINQFNKK255 pKa = 10.44GNLGNRR261 pKa = 11.84FSIPIPSVSRR271 pKa = 11.84PSSLTKK277 pKa = 10.3QVTINPKK284 pKa = 8.47QQ285 pKa = 3.09
Molecular weight: 32.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1266 |
285 |
527 |
422.0 |
47.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.687 ± 1.086 | 1.343 ± 0.621 |
4.265 ± 0.323 | 5.134 ± 1.281 |
4.344 ± 0.375 | 4.66 ± 0.65 |
1.896 ± 0.597 | 6.951 ± 0.526 |
6.319 ± 0.873 | 8.768 ± 0.957 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.449 ± 0.652 | 6.872 ± 0.367 |
5.766 ± 0.479 | 4.028 ± 0.262 |
5.134 ± 1.047 | 7.899 ± 1.331 |
7.82 ± 1.971 | 5.687 ± 0.597 |
0.79 ± 0.156 | 4.186 ± 0.44 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
