
MSSI2.225 virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus humas1; Human associated gemykibivirus 1
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
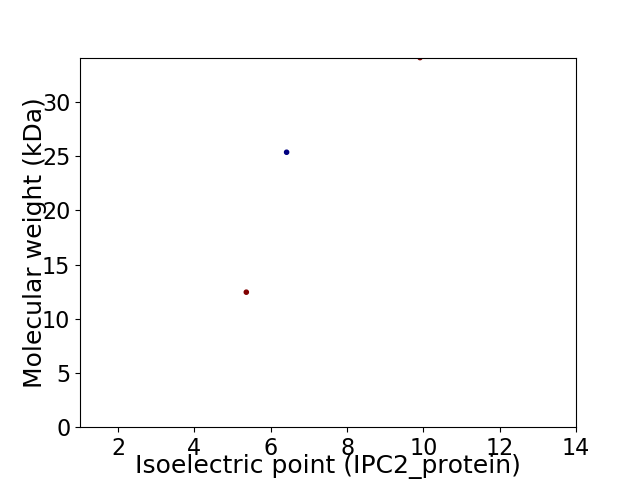
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
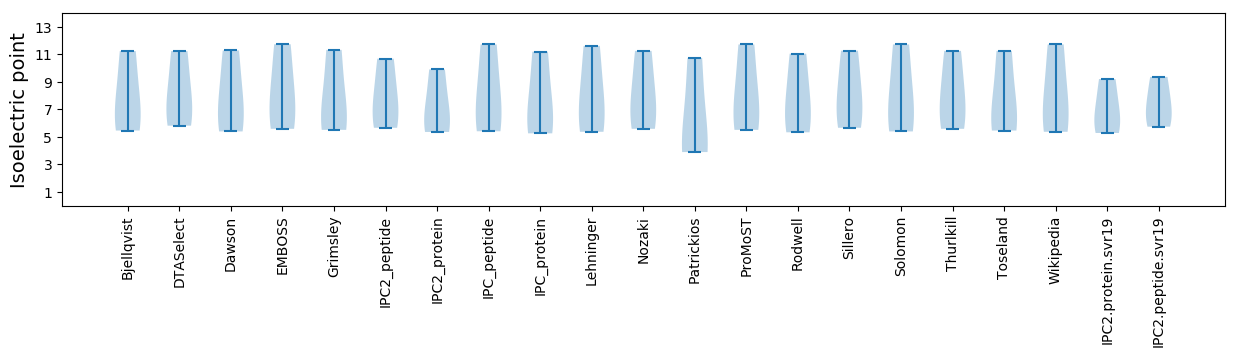
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A077XLY5|A0A077XLY5_9VIRU Replication associated protein OS=MSSI2.225 virus OX=1516081 GN=rep PE=4 SV=1
MM1 pKa = 7.68GKK3 pKa = 8.35TIWARR8 pKa = 11.84SLGNHH13 pKa = 7.12AYY15 pKa = 10.41FGGLFSLDD23 pKa = 3.56EE24 pKa = 4.33NLDD27 pKa = 3.61GVEE30 pKa = 3.93YY31 pKa = 10.64AIFDD35 pKa = 3.87DD36 pKa = 4.21FGGIKK41 pKa = 10.32FLPSYY46 pKa = 10.27KK47 pKa = 10.0FWLGHH52 pKa = 4.73QKK54 pKa = 9.95QFYY57 pKa = 8.86VTDD60 pKa = 3.67KK61 pKa = 11.21YY62 pKa = 10.94KK63 pKa = 10.96GKK65 pKa = 10.68KK66 pKa = 8.0LVHH69 pKa = 6.36WARR72 pKa = 11.84PSIWLSNSDD81 pKa = 3.6PRR83 pKa = 11.84DD84 pKa = 3.43EE85 pKa = 5.63LGVDD89 pKa = 4.0TEE91 pKa = 4.54WLNANCDD98 pKa = 3.81FVYY101 pKa = 10.57LDD103 pKa = 4.01SPIVSS108 pKa = 3.68
MM1 pKa = 7.68GKK3 pKa = 8.35TIWARR8 pKa = 11.84SLGNHH13 pKa = 7.12AYY15 pKa = 10.41FGGLFSLDD23 pKa = 3.56EE24 pKa = 4.33NLDD27 pKa = 3.61GVEE30 pKa = 3.93YY31 pKa = 10.64AIFDD35 pKa = 3.87DD36 pKa = 4.21FGGIKK41 pKa = 10.32FLPSYY46 pKa = 10.27KK47 pKa = 10.0FWLGHH52 pKa = 4.73QKK54 pKa = 9.95QFYY57 pKa = 8.86VTDD60 pKa = 3.67KK61 pKa = 11.21YY62 pKa = 10.94KK63 pKa = 10.96GKK65 pKa = 10.68KK66 pKa = 8.0LVHH69 pKa = 6.36WARR72 pKa = 11.84PSIWLSNSDD81 pKa = 3.6PRR83 pKa = 11.84DD84 pKa = 3.43EE85 pKa = 5.63LGVDD89 pKa = 4.0TEE91 pKa = 4.54WLNANCDD98 pKa = 3.81FVYY101 pKa = 10.57LDD103 pKa = 4.01SPIVSS108 pKa = 3.68
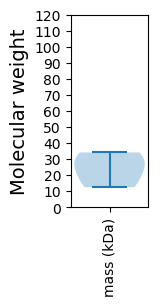
Molecular weight: 12.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077XL96|A0A077XL96_9VIRU Replication protein OS=MSSI2.225 virus OX=1516081 GN=Rep PE=3 SV=1
MM1 pKa = 7.18SVARR5 pKa = 11.84RR6 pKa = 11.84SGRR9 pKa = 11.84RR10 pKa = 11.84TRR12 pKa = 11.84KK13 pKa = 8.7AFRR16 pKa = 11.84RR17 pKa = 11.84GGRR20 pKa = 11.84ATRR23 pKa = 11.84SRR25 pKa = 11.84GRR27 pKa = 11.84STRR30 pKa = 11.84RR31 pKa = 11.84SSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84TMTTRR45 pKa = 11.84RR46 pKa = 11.84VLNITTRR53 pKa = 11.84KK54 pKa = 9.91KK55 pKa = 9.8VDD57 pKa = 2.92NMLPVVVAEE66 pKa = 4.85DD67 pKa = 3.37NTTTVGPYY75 pKa = 9.82TSVSPLLSLFVPNARR90 pKa = 11.84TTRR93 pKa = 11.84TPVTNPAVRR102 pKa = 11.84NSSDD106 pKa = 2.71IFAVGYY112 pKa = 8.9RR113 pKa = 11.84EE114 pKa = 3.91RR115 pKa = 11.84VRR117 pKa = 11.84IDD119 pKa = 3.2LMGGGTFMWRR129 pKa = 11.84RR130 pKa = 11.84IVFMLKK136 pKa = 10.54GSDD139 pKa = 3.54LRR141 pKa = 11.84VAMNSSDD148 pKa = 3.6SGNIPNQLFDD158 pKa = 3.24QTTEE162 pKa = 4.13GGCRR166 pKa = 11.84RR167 pKa = 11.84VIGPLLGVTNARR179 pKa = 11.84DD180 pKa = 3.49EE181 pKa = 4.29LQSYY185 pKa = 7.08VFRR188 pKa = 11.84GQEE191 pKa = 3.7DD192 pKa = 4.19SDD194 pKa = 3.57WADD197 pKa = 3.23QFTAPLDD204 pKa = 3.63TRR206 pKa = 11.84RR207 pKa = 11.84ITVKK211 pKa = 10.22SDD213 pKa = 2.63KK214 pKa = 10.77LRR216 pKa = 11.84VIRR219 pKa = 11.84PGNEE223 pKa = 3.34TGASRR228 pKa = 11.84LFKK231 pKa = 9.76FWYY234 pKa = 8.43PIRR237 pKa = 11.84RR238 pKa = 11.84TISYY242 pKa = 10.58EE243 pKa = 3.91DD244 pKa = 4.05DD245 pKa = 3.97LEE247 pKa = 6.14SDD249 pKa = 4.19VVGDD253 pKa = 4.43RR254 pKa = 11.84PFSTAGLRR262 pKa = 11.84GVGDD266 pKa = 4.13MYY268 pKa = 11.73VMDD271 pKa = 4.6IMGITNLVNDD281 pKa = 5.3APPTSYY287 pKa = 10.97RR288 pKa = 11.84FGPEE292 pKa = 3.53GSFYY296 pKa = 9.46WHH298 pKa = 6.64EE299 pKa = 4.04RR300 pKa = 3.36
MM1 pKa = 7.18SVARR5 pKa = 11.84RR6 pKa = 11.84SGRR9 pKa = 11.84RR10 pKa = 11.84TRR12 pKa = 11.84KK13 pKa = 8.7AFRR16 pKa = 11.84RR17 pKa = 11.84GGRR20 pKa = 11.84ATRR23 pKa = 11.84SRR25 pKa = 11.84GRR27 pKa = 11.84STRR30 pKa = 11.84RR31 pKa = 11.84SSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84TMTTRR45 pKa = 11.84RR46 pKa = 11.84VLNITTRR53 pKa = 11.84KK54 pKa = 9.91KK55 pKa = 9.8VDD57 pKa = 2.92NMLPVVVAEE66 pKa = 4.85DD67 pKa = 3.37NTTTVGPYY75 pKa = 9.82TSVSPLLSLFVPNARR90 pKa = 11.84TTRR93 pKa = 11.84TPVTNPAVRR102 pKa = 11.84NSSDD106 pKa = 2.71IFAVGYY112 pKa = 8.9RR113 pKa = 11.84EE114 pKa = 3.91RR115 pKa = 11.84VRR117 pKa = 11.84IDD119 pKa = 3.2LMGGGTFMWRR129 pKa = 11.84RR130 pKa = 11.84IVFMLKK136 pKa = 10.54GSDD139 pKa = 3.54LRR141 pKa = 11.84VAMNSSDD148 pKa = 3.6SGNIPNQLFDD158 pKa = 3.24QTTEE162 pKa = 4.13GGCRR166 pKa = 11.84RR167 pKa = 11.84VIGPLLGVTNARR179 pKa = 11.84DD180 pKa = 3.49EE181 pKa = 4.29LQSYY185 pKa = 7.08VFRR188 pKa = 11.84GQEE191 pKa = 3.7DD192 pKa = 4.19SDD194 pKa = 3.57WADD197 pKa = 3.23QFTAPLDD204 pKa = 3.63TRR206 pKa = 11.84RR207 pKa = 11.84ITVKK211 pKa = 10.22SDD213 pKa = 2.63KK214 pKa = 10.77LRR216 pKa = 11.84VIRR219 pKa = 11.84PGNEE223 pKa = 3.34TGASRR228 pKa = 11.84LFKK231 pKa = 9.76FWYY234 pKa = 8.43PIRR237 pKa = 11.84RR238 pKa = 11.84TISYY242 pKa = 10.58EE243 pKa = 3.91DD244 pKa = 4.05DD245 pKa = 3.97LEE247 pKa = 6.14SDD249 pKa = 4.19VVGDD253 pKa = 4.43RR254 pKa = 11.84PFSTAGLRR262 pKa = 11.84GVGDD266 pKa = 4.13MYY268 pKa = 11.73VMDD271 pKa = 4.6IMGITNLVNDD281 pKa = 5.3APPTSYY287 pKa = 10.97RR288 pKa = 11.84FGPEE292 pKa = 3.53GSFYY296 pKa = 9.46WHH298 pKa = 6.64EE299 pKa = 4.04RR300 pKa = 3.36
Molecular weight: 34.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
640 |
108 |
300 |
213.3 |
23.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.875 ± 1.799 | 1.25 ± 0.646 |
6.406 ± 0.885 | 4.375 ± 0.78 |
4.844 ± 0.671 | 9.219 ± 0.581 |
1.406 ± 0.666 | 3.906 ± 0.235 |
3.281 ± 1.091 | 7.5 ± 0.992 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.031 ± 0.796 | 4.219 ± 0.161 |
4.688 ± 0.276 | 2.656 ± 0.77 |
10.625 ± 3.054 | 7.187 ± 0.564 |
6.563 ± 1.732 | 7.656 ± 0.704 |
2.188 ± 0.705 | 3.125 ± 0.635 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
