
Streptomyces phage Toma
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Arquatrovirinae; Likavirus; unclassified Likavirus
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
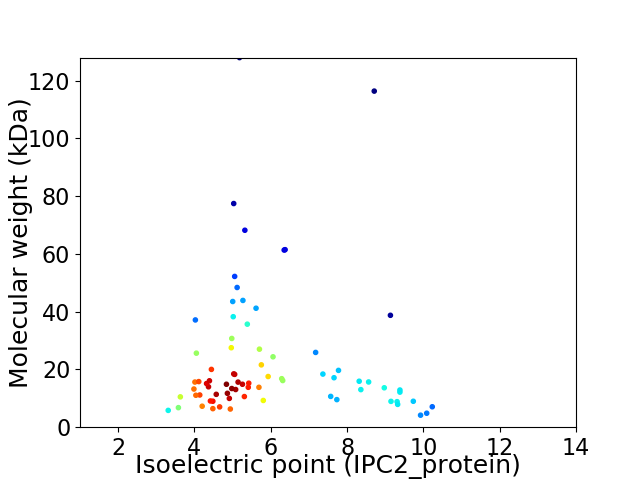
Virtual 2D-PAGE plot for 76 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U8UUB6|A0A2U8UUB6_9CAUD Uncharacterized protein OS=Streptomyces phage Toma OX=2182323 GN=69 PE=4 SV=1
MM1 pKa = 7.4IGGVVRR7 pKa = 11.84GSDD10 pKa = 3.78FRR12 pKa = 11.84CWTCVDD18 pKa = 4.41FSSYY22 pKa = 9.55EE23 pKa = 3.99RR24 pKa = 11.84EE25 pKa = 4.38TNSGDD30 pKa = 3.43AAEE33 pKa = 4.48KK34 pKa = 9.54CTYY37 pKa = 10.36CKK39 pKa = 10.73GPVWSQEE46 pKa = 3.84EE47 pKa = 4.33TTVNVDD53 pKa = 2.25IDD55 pKa = 4.27TVQEE59 pKa = 4.09YY60 pKa = 11.2ADD62 pKa = 3.75NALEE66 pKa = 4.36RR67 pKa = 11.84PSDD70 pKa = 3.32AAFWDD75 pKa = 3.94DD76 pKa = 3.64RR77 pKa = 11.84CYY79 pKa = 8.14TTHH82 pKa = 7.01APVIGWADD90 pKa = 3.62RR91 pKa = 11.84GDD93 pKa = 4.95DD94 pKa = 3.38ILEE97 pKa = 4.06EE98 pKa = 4.48SNYY101 pKa = 10.22HH102 pKa = 4.97SAKK105 pKa = 10.73ALIEE109 pKa = 4.29GAATEE114 pKa = 4.46DD115 pKa = 3.28EE116 pKa = 4.67HH117 pKa = 8.99VFEE120 pKa = 4.88GTAGHH125 pKa = 6.71WLVGSLSQVWVQVYY139 pKa = 7.0EE140 pKa = 4.13TRR142 pKa = 11.84PEE144 pKa = 4.09CSTIGCEE151 pKa = 4.46EE152 pKa = 3.96EE153 pKa = 4.18SEE155 pKa = 4.3YY156 pKa = 11.22LATYY160 pKa = 8.87TLASYY165 pKa = 10.76DD166 pKa = 3.71SEE168 pKa = 4.71GFCAEE173 pKa = 4.13HH174 pKa = 7.35KK175 pKa = 9.46EE176 pKa = 4.05ALEE179 pKa = 4.22GAFLAEE185 pKa = 4.15LLRR188 pKa = 11.84IEE190 pKa = 5.38FEE192 pKa = 3.85DD193 pKa = 3.72HH194 pKa = 6.89PRR196 pKa = 11.84DD197 pKa = 3.49FTAAFIEE204 pKa = 4.27AVEE207 pKa = 4.07IQEE210 pKa = 4.21ALKK213 pKa = 10.42DD214 pKa = 3.96YY215 pKa = 10.28PILDD219 pKa = 3.47EE220 pKa = 4.27TDD222 pKa = 3.43FSEE225 pKa = 5.25RR226 pKa = 11.84EE227 pKa = 3.77WEE229 pKa = 4.01RR230 pKa = 11.84FEE232 pKa = 5.12EE233 pKa = 4.14NSKK236 pKa = 10.6EE237 pKa = 4.19AIEE240 pKa = 4.2EE241 pKa = 4.11AQRR244 pKa = 11.84SHH246 pKa = 7.16PEE248 pKa = 3.79DD249 pKa = 4.05SIEE252 pKa = 5.6DD253 pKa = 3.19ITEE256 pKa = 3.58IEE258 pKa = 3.79NRR260 pKa = 11.84IFSEE264 pKa = 4.31SALSDD269 pKa = 3.4LFGYY273 pKa = 9.58EE274 pKa = 4.0GNAEE278 pKa = 4.19VSWDD282 pKa = 3.82AVADD286 pKa = 3.73IYY288 pKa = 11.6DD289 pKa = 4.23EE290 pKa = 4.37YY291 pKa = 11.39RR292 pKa = 11.84DD293 pKa = 3.69AYY295 pKa = 9.87FLEE298 pKa = 4.93RR299 pKa = 11.84ATEE302 pKa = 4.26VYY304 pKa = 9.2RR305 pKa = 11.84WNYY308 pKa = 10.13LGYY311 pKa = 10.9NPDD314 pKa = 3.4QLEE317 pKa = 4.25LFVIIIAAA325 pKa = 3.66
MM1 pKa = 7.4IGGVVRR7 pKa = 11.84GSDD10 pKa = 3.78FRR12 pKa = 11.84CWTCVDD18 pKa = 4.41FSSYY22 pKa = 9.55EE23 pKa = 3.99RR24 pKa = 11.84EE25 pKa = 4.38TNSGDD30 pKa = 3.43AAEE33 pKa = 4.48KK34 pKa = 9.54CTYY37 pKa = 10.36CKK39 pKa = 10.73GPVWSQEE46 pKa = 3.84EE47 pKa = 4.33TTVNVDD53 pKa = 2.25IDD55 pKa = 4.27TVQEE59 pKa = 4.09YY60 pKa = 11.2ADD62 pKa = 3.75NALEE66 pKa = 4.36RR67 pKa = 11.84PSDD70 pKa = 3.32AAFWDD75 pKa = 3.94DD76 pKa = 3.64RR77 pKa = 11.84CYY79 pKa = 8.14TTHH82 pKa = 7.01APVIGWADD90 pKa = 3.62RR91 pKa = 11.84GDD93 pKa = 4.95DD94 pKa = 3.38ILEE97 pKa = 4.06EE98 pKa = 4.48SNYY101 pKa = 10.22HH102 pKa = 4.97SAKK105 pKa = 10.73ALIEE109 pKa = 4.29GAATEE114 pKa = 4.46DD115 pKa = 3.28EE116 pKa = 4.67HH117 pKa = 8.99VFEE120 pKa = 4.88GTAGHH125 pKa = 6.71WLVGSLSQVWVQVYY139 pKa = 7.0EE140 pKa = 4.13TRR142 pKa = 11.84PEE144 pKa = 4.09CSTIGCEE151 pKa = 4.46EE152 pKa = 3.96EE153 pKa = 4.18SEE155 pKa = 4.3YY156 pKa = 11.22LATYY160 pKa = 8.87TLASYY165 pKa = 10.76DD166 pKa = 3.71SEE168 pKa = 4.71GFCAEE173 pKa = 4.13HH174 pKa = 7.35KK175 pKa = 9.46EE176 pKa = 4.05ALEE179 pKa = 4.22GAFLAEE185 pKa = 4.15LLRR188 pKa = 11.84IEE190 pKa = 5.38FEE192 pKa = 3.85DD193 pKa = 3.72HH194 pKa = 6.89PRR196 pKa = 11.84DD197 pKa = 3.49FTAAFIEE204 pKa = 4.27AVEE207 pKa = 4.07IQEE210 pKa = 4.21ALKK213 pKa = 10.42DD214 pKa = 3.96YY215 pKa = 10.28PILDD219 pKa = 3.47EE220 pKa = 4.27TDD222 pKa = 3.43FSEE225 pKa = 5.25RR226 pKa = 11.84EE227 pKa = 3.77WEE229 pKa = 4.01RR230 pKa = 11.84FEE232 pKa = 5.12EE233 pKa = 4.14NSKK236 pKa = 10.6EE237 pKa = 4.19AIEE240 pKa = 4.2EE241 pKa = 4.11AQRR244 pKa = 11.84SHH246 pKa = 7.16PEE248 pKa = 3.79DD249 pKa = 4.05SIEE252 pKa = 5.6DD253 pKa = 3.19ITEE256 pKa = 3.58IEE258 pKa = 3.79NRR260 pKa = 11.84IFSEE264 pKa = 4.31SALSDD269 pKa = 3.4LFGYY273 pKa = 9.58EE274 pKa = 4.0GNAEE278 pKa = 4.19VSWDD282 pKa = 3.82AVADD286 pKa = 3.73IYY288 pKa = 11.6DD289 pKa = 4.23EE290 pKa = 4.37YY291 pKa = 11.39RR292 pKa = 11.84DD293 pKa = 3.69AYY295 pKa = 9.87FLEE298 pKa = 4.93RR299 pKa = 11.84ATEE302 pKa = 4.26VYY304 pKa = 9.2RR305 pKa = 11.84WNYY308 pKa = 10.13LGYY311 pKa = 10.9NPDD314 pKa = 3.4QLEE317 pKa = 4.25LFVIIIAAA325 pKa = 3.66
Molecular weight: 37.13 kDa
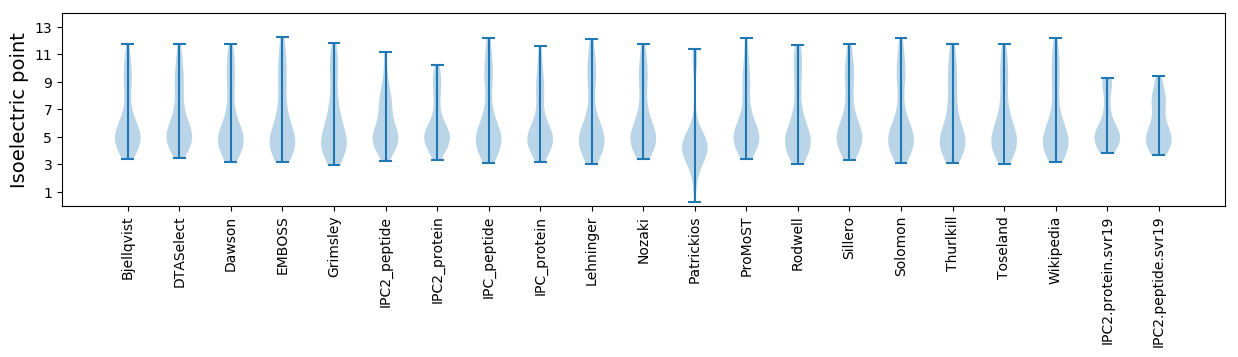
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U8UU58|A0A2U8UU58_9CAUD Uncharacterized protein OS=Streptomyces phage Toma OX=2182323 GN=65 PE=4 SV=1
MM1 pKa = 7.5AGWSGSDD8 pKa = 2.78RR9 pKa = 11.84RR10 pKa = 11.84QRR12 pKa = 11.84LPQNWAQIRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84ILRR26 pKa = 11.84RR27 pKa = 11.84DD28 pKa = 3.24GNRR31 pKa = 11.84CVWVFEE37 pKa = 4.3GKK39 pKa = 9.93RR40 pKa = 11.84CEE42 pKa = 4.2EE43 pKa = 4.1VATEE47 pKa = 4.03VDD49 pKa = 4.17HH50 pKa = 7.66IIAGDD55 pKa = 3.55NHH57 pKa = 7.38DD58 pKa = 4.65DD59 pKa = 3.68SNLRR63 pKa = 11.84SLCSWHH69 pKa = 6.31HH70 pKa = 4.87QKK72 pKa = 10.65KK73 pKa = 10.28SSSEE77 pKa = 3.48GRR79 pKa = 11.84AAQLAKK85 pKa = 10.19RR86 pKa = 11.84RR87 pKa = 11.84QIEE90 pKa = 3.73KK91 pKa = 9.69RR92 pKa = 11.84FRR94 pKa = 11.84RR95 pKa = 11.84NEE97 pKa = 3.86SHH99 pKa = 7.53PGLLL103 pKa = 4.09
MM1 pKa = 7.5AGWSGSDD8 pKa = 2.78RR9 pKa = 11.84RR10 pKa = 11.84QRR12 pKa = 11.84LPQNWAQIRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84ILRR26 pKa = 11.84RR27 pKa = 11.84DD28 pKa = 3.24GNRR31 pKa = 11.84CVWVFEE37 pKa = 4.3GKK39 pKa = 9.93RR40 pKa = 11.84CEE42 pKa = 4.2EE43 pKa = 4.1VATEE47 pKa = 4.03VDD49 pKa = 4.17HH50 pKa = 7.66IIAGDD55 pKa = 3.55NHH57 pKa = 7.38DD58 pKa = 4.65DD59 pKa = 3.68SNLRR63 pKa = 11.84SLCSWHH69 pKa = 6.31HH70 pKa = 4.87QKK72 pKa = 10.65KK73 pKa = 10.28SSSEE77 pKa = 3.48GRR79 pKa = 11.84AAQLAKK85 pKa = 10.19RR86 pKa = 11.84RR87 pKa = 11.84QIEE90 pKa = 3.73KK91 pKa = 9.69RR92 pKa = 11.84FRR94 pKa = 11.84RR95 pKa = 11.84NEE97 pKa = 3.86SHH99 pKa = 7.53PGLLL103 pKa = 4.09
Molecular weight: 12.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
15820 |
39 |
1160 |
208.2 |
22.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.335 ± 0.376 | 0.721 ± 0.105 |
6.403 ± 0.228 | 7.143 ± 0.468 |
3.198 ± 0.154 | 8.666 ± 0.431 |
2.042 ± 0.184 | 4.576 ± 0.37 |
4.469 ± 0.225 | 8.407 ± 0.418 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.212 ± 0.162 | 3.104 ± 0.151 |
4.829 ± 0.19 | 3.274 ± 0.159 |
6.391 ± 0.398 | 5.885 ± 0.275 |
6.391 ± 0.342 | 7.149 ± 0.252 |
1.934 ± 0.121 | 2.87 ± 0.276 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
