
Microbacterium sp. dk485
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
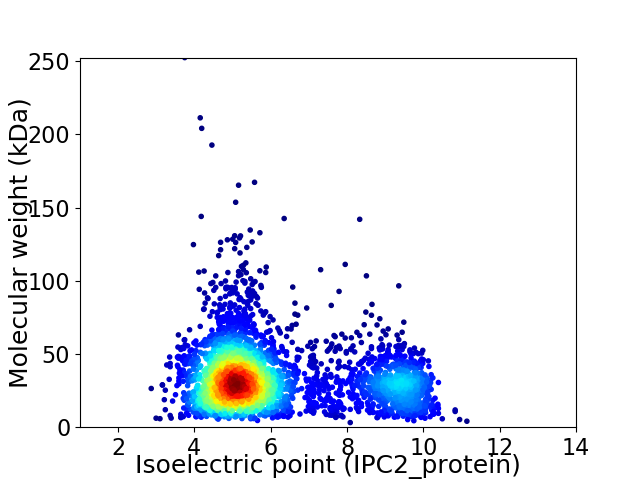
Virtual 2D-PAGE plot for 3585 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
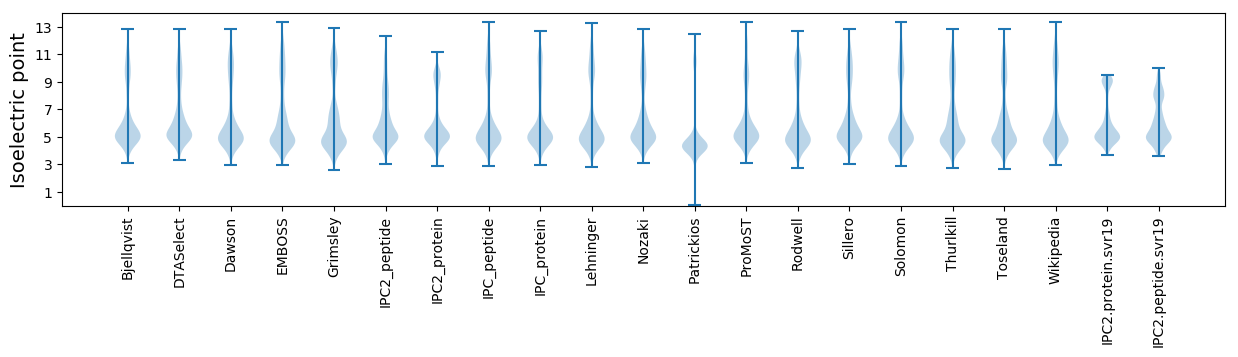
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y9PK63|A0A4Y9PK63_9MICO LysE family translocator OS=Microbacterium sp. dk485 OX=2560021 GN=E4V99_17155 PE=4 SV=1
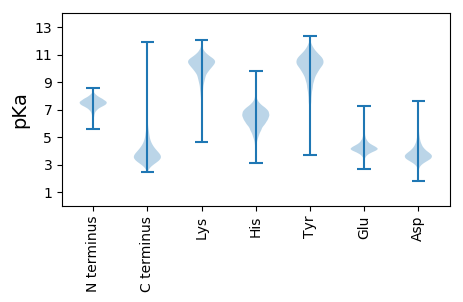
MM1 pKa = 7.46SGAALAVTALLVAGCSAGEE20 pKa = 4.12GVDD23 pKa = 4.3LDD25 pKa = 3.85SSGADD30 pKa = 3.32GEE32 pKa = 4.73VLVAAIGGEE41 pKa = 4.05PDD43 pKa = 3.24QLDD46 pKa = 3.79PQKK49 pKa = 8.03TTSYY53 pKa = 10.51FAFEE57 pKa = 3.92VLEE60 pKa = 4.21NVFDD64 pKa = 4.06TLVEE68 pKa = 4.24PDD70 pKa = 4.9EE71 pKa = 4.24NLQMQPALAEE81 pKa = 4.09SWEE84 pKa = 4.14VSDD87 pKa = 6.39DD88 pKa = 3.6QLEE91 pKa = 4.38WTFTLRR97 pKa = 11.84DD98 pKa = 3.35GVTFHH103 pKa = 7.53DD104 pKa = 4.55GSEE107 pKa = 4.21FTSADD112 pKa = 2.79VMYY115 pKa = 10.28SYY117 pKa = 11.53DD118 pKa = 4.08RR119 pKa = 11.84IIDD122 pKa = 3.92EE123 pKa = 4.56EE124 pKa = 4.85LSNAWKK130 pKa = 9.4FASVTDD136 pKa = 3.42VTAPDD141 pKa = 3.91DD142 pKa = 3.66QTVVITVAQPTPNLLSNLGGFKK164 pKa = 10.63GMAIVQEE171 pKa = 4.51DD172 pKa = 3.87NVASGDD178 pKa = 3.32ITTAPVGTGPFSVEE192 pKa = 4.01EE193 pKa = 4.33YY194 pKa = 10.93VSGDD198 pKa = 4.04HH199 pKa = 6.45ITLTANPDD207 pKa = 3.26YY208 pKa = 10.71WGGAPEE214 pKa = 4.45LGGVEE219 pKa = 3.94YY220 pKa = 10.86RR221 pKa = 11.84FISEE225 pKa = 4.15PATALAALRR234 pKa = 11.84GGEE237 pKa = 4.14IQWTDD242 pKa = 3.26VVPPQQVGEE251 pKa = 4.4LEE253 pKa = 3.92TDD255 pKa = 3.18EE256 pKa = 5.15ALEE259 pKa = 4.46LGVTPSSDD267 pKa = 3.04YY268 pKa = 10.62WYY270 pKa = 10.62LALNEE275 pKa = 4.21ARR277 pKa = 11.84EE278 pKa = 4.18PWSDD282 pKa = 3.19VRR284 pKa = 11.84VRR286 pKa = 11.84QAIAYY291 pKa = 9.59AIDD294 pKa = 3.59RR295 pKa = 11.84DD296 pKa = 4.36AIVQAVSYY304 pKa = 8.07GTAEE308 pKa = 4.2LNQLAIPEE316 pKa = 4.01QSVWYY321 pKa = 8.39TPYY324 pKa = 9.85DD325 pKa = 3.68TYY327 pKa = 11.13STDD330 pKa = 3.15MDD332 pKa = 4.07TAQQLMAEE340 pKa = 4.34AGFTGGTLDD349 pKa = 5.27LLATSDD355 pKa = 3.86YY356 pKa = 10.71PEE358 pKa = 4.57TVTAAQIIAANLEE371 pKa = 3.98PLGIEE376 pKa = 4.22VAIRR380 pKa = 11.84QPDD383 pKa = 4.05FSTWLDD389 pKa = 3.61EE390 pKa = 4.3QNSGNFDD397 pKa = 3.71MLMMGWLGNIDD408 pKa = 4.23PDD410 pKa = 3.72DD411 pKa = 4.38FYY413 pKa = 11.75YY414 pKa = 10.64SQHH417 pKa = 6.51HH418 pKa = 5.6STGASNAQKK427 pKa = 10.3YY428 pKa = 9.85ANPEE432 pKa = 3.45VDD434 pKa = 3.3QLLDD438 pKa = 3.52AGRR441 pKa = 11.84VEE443 pKa = 4.56TDD445 pKa = 2.89TEE447 pKa = 4.08ARR449 pKa = 11.84KK450 pKa = 9.99DD451 pKa = 3.77LYY453 pKa = 11.11AQAATIIADD462 pKa = 3.73EE463 pKa = 4.11ASYY466 pKa = 10.53IYY468 pKa = 10.34LYY470 pKa = 10.51NPSVIQAWSPSITGYY485 pKa = 8.02EE486 pKa = 3.73ARR488 pKa = 11.84ADD490 pKa = 3.79RR491 pKa = 11.84AIRR494 pKa = 11.84FRR496 pKa = 11.84DD497 pKa = 3.38AALSGG502 pKa = 3.56
MM1 pKa = 7.46SGAALAVTALLVAGCSAGEE20 pKa = 4.12GVDD23 pKa = 4.3LDD25 pKa = 3.85SSGADD30 pKa = 3.32GEE32 pKa = 4.73VLVAAIGGEE41 pKa = 4.05PDD43 pKa = 3.24QLDD46 pKa = 3.79PQKK49 pKa = 8.03TTSYY53 pKa = 10.51FAFEE57 pKa = 3.92VLEE60 pKa = 4.21NVFDD64 pKa = 4.06TLVEE68 pKa = 4.24PDD70 pKa = 4.9EE71 pKa = 4.24NLQMQPALAEE81 pKa = 4.09SWEE84 pKa = 4.14VSDD87 pKa = 6.39DD88 pKa = 3.6QLEE91 pKa = 4.38WTFTLRR97 pKa = 11.84DD98 pKa = 3.35GVTFHH103 pKa = 7.53DD104 pKa = 4.55GSEE107 pKa = 4.21FTSADD112 pKa = 2.79VMYY115 pKa = 10.28SYY117 pKa = 11.53DD118 pKa = 4.08RR119 pKa = 11.84IIDD122 pKa = 3.92EE123 pKa = 4.56EE124 pKa = 4.85LSNAWKK130 pKa = 9.4FASVTDD136 pKa = 3.42VTAPDD141 pKa = 3.91DD142 pKa = 3.66QTVVITVAQPTPNLLSNLGGFKK164 pKa = 10.63GMAIVQEE171 pKa = 4.51DD172 pKa = 3.87NVASGDD178 pKa = 3.32ITTAPVGTGPFSVEE192 pKa = 4.01EE193 pKa = 4.33YY194 pKa = 10.93VSGDD198 pKa = 4.04HH199 pKa = 6.45ITLTANPDD207 pKa = 3.26YY208 pKa = 10.71WGGAPEE214 pKa = 4.45LGGVEE219 pKa = 3.94YY220 pKa = 10.86RR221 pKa = 11.84FISEE225 pKa = 4.15PATALAALRR234 pKa = 11.84GGEE237 pKa = 4.14IQWTDD242 pKa = 3.26VVPPQQVGEE251 pKa = 4.4LEE253 pKa = 3.92TDD255 pKa = 3.18EE256 pKa = 5.15ALEE259 pKa = 4.46LGVTPSSDD267 pKa = 3.04YY268 pKa = 10.62WYY270 pKa = 10.62LALNEE275 pKa = 4.21ARR277 pKa = 11.84EE278 pKa = 4.18PWSDD282 pKa = 3.19VRR284 pKa = 11.84VRR286 pKa = 11.84QAIAYY291 pKa = 9.59AIDD294 pKa = 3.59RR295 pKa = 11.84DD296 pKa = 4.36AIVQAVSYY304 pKa = 8.07GTAEE308 pKa = 4.2LNQLAIPEE316 pKa = 4.01QSVWYY321 pKa = 8.39TPYY324 pKa = 9.85DD325 pKa = 3.68TYY327 pKa = 11.13STDD330 pKa = 3.15MDD332 pKa = 4.07TAQQLMAEE340 pKa = 4.34AGFTGGTLDD349 pKa = 5.27LLATSDD355 pKa = 3.86YY356 pKa = 10.71PEE358 pKa = 4.57TVTAAQIIAANLEE371 pKa = 3.98PLGIEE376 pKa = 4.22VAIRR380 pKa = 11.84QPDD383 pKa = 4.05FSTWLDD389 pKa = 3.61EE390 pKa = 4.3QNSGNFDD397 pKa = 3.71MLMMGWLGNIDD408 pKa = 4.23PDD410 pKa = 3.72DD411 pKa = 4.38FYY413 pKa = 11.75YY414 pKa = 10.64SQHH417 pKa = 6.51HH418 pKa = 5.6STGASNAQKK427 pKa = 10.3YY428 pKa = 9.85ANPEE432 pKa = 3.45VDD434 pKa = 3.3QLLDD438 pKa = 3.52AGRR441 pKa = 11.84VEE443 pKa = 4.56TDD445 pKa = 2.89TEE447 pKa = 4.08ARR449 pKa = 11.84KK450 pKa = 9.99DD451 pKa = 3.77LYY453 pKa = 11.11AQAATIIADD462 pKa = 3.73EE463 pKa = 4.11ASYY466 pKa = 10.53IYY468 pKa = 10.34LYY470 pKa = 10.51NPSVIQAWSPSITGYY485 pKa = 8.02EE486 pKa = 3.73ARR488 pKa = 11.84ADD490 pKa = 3.79RR491 pKa = 11.84AIRR494 pKa = 11.84FRR496 pKa = 11.84DD497 pKa = 3.38AALSGG502 pKa = 3.56
Molecular weight: 54.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9PXU4|A0A4Y9PXU4_9MICO Phenylacetate-CoA oxygenase subunit PaaJ OS=Microbacterium sp. dk485 OX=2560021 GN=paaJ PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1157956 |
29 |
2480 |
323.0 |
34.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.219 ± 0.066 | 0.49 ± 0.009 |
6.223 ± 0.037 | 5.615 ± 0.038 |
3.072 ± 0.026 | 9.034 ± 0.035 |
1.976 ± 0.022 | 4.237 ± 0.032 |
1.541 ± 0.03 | 10.165 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.765 ± 0.015 | 1.729 ± 0.02 |
5.7 ± 0.031 | 2.65 ± 0.019 |
7.759 ± 0.048 | 5.293 ± 0.026 |
5.852 ± 0.032 | 9.16 ± 0.035 |
1.564 ± 0.019 | 1.955 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
