
Aliifodinibius sediminis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Balneolaeota; Balneolia; Balneolales; Balneolaceae; Aliifodinibius
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
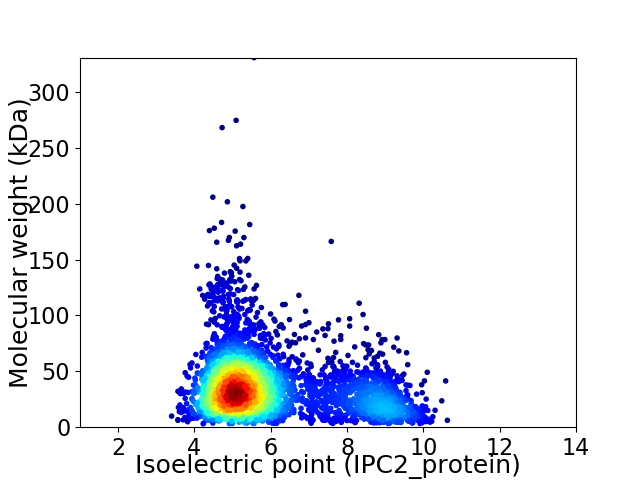
Virtual 2D-PAGE plot for 3822 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A521B7J8|A0A521B7J8_9BACT Uncharacterized protein OS=Aliifodinibius sediminis OX=1214077 GN=SAMN06265218_102263 PE=4 SV=1
MM1 pKa = 7.32QSIVNNRR8 pKa = 11.84FCLLPVILLISSLALTSCLDD28 pKa = 3.2SSTDD32 pKa = 3.53TNQGASKK39 pKa = 10.79NLLEE43 pKa = 4.26QAQSYY48 pKa = 11.42ADD50 pKa = 3.71FDD52 pKa = 4.27TFVDD56 pKa = 4.73IISDD60 pKa = 3.91TEE62 pKa = 4.42LNTTIANEE70 pKa = 4.51GPLTLLVPTNDD81 pKa = 4.11AFDD84 pKa = 4.6ALPSGTLEE92 pKa = 4.34SLTDD96 pKa = 3.52EE97 pKa = 4.08QLIDD101 pKa = 3.21ILSYY105 pKa = 10.79HH106 pKa = 6.82VIEE109 pKa = 5.59QIIDD113 pKa = 3.78LNQLSAEE120 pKa = 4.18EE121 pKa = 5.03TFTSMQGDD129 pKa = 3.6NIFFVISNDD138 pKa = 3.01SLYY141 pKa = 10.49INGSNYY147 pKa = 10.08VGGVGASNGVLYY159 pKa = 9.01ATDD162 pKa = 3.27QVLFPDD168 pKa = 4.78AYY170 pKa = 11.24LDD172 pKa = 3.75VTGLVHH178 pKa = 7.42KK179 pKa = 10.44RR180 pKa = 11.84KK181 pKa = 9.35QLSEE185 pKa = 4.05LDD187 pKa = 3.55KK188 pKa = 11.54SIDD191 pKa = 3.6NTGLASTLEE200 pKa = 4.48DD201 pKa = 3.05TSSAYY206 pKa = 9.75TIFAPSNTALQNAEE220 pKa = 4.39LSADD224 pKa = 3.5TLKK227 pKa = 10.98YY228 pKa = 10.24HH229 pKa = 5.65VVSGKK234 pKa = 10.21LLSQDD239 pKa = 3.62FSSSQTYY246 pKa = 6.76TTLNGKK252 pKa = 8.02EE253 pKa = 4.06LSVEE257 pKa = 4.11VSGSSVTINGEE268 pKa = 4.06AAVTTADD275 pKa = 3.22IEE277 pKa = 4.61GVNGVVHH284 pKa = 7.33IIDD287 pKa = 3.66TALAPPSEE295 pKa = 4.32
MM1 pKa = 7.32QSIVNNRR8 pKa = 11.84FCLLPVILLISSLALTSCLDD28 pKa = 3.2SSTDD32 pKa = 3.53TNQGASKK39 pKa = 10.79NLLEE43 pKa = 4.26QAQSYY48 pKa = 11.42ADD50 pKa = 3.71FDD52 pKa = 4.27TFVDD56 pKa = 4.73IISDD60 pKa = 3.91TEE62 pKa = 4.42LNTTIANEE70 pKa = 4.51GPLTLLVPTNDD81 pKa = 4.11AFDD84 pKa = 4.6ALPSGTLEE92 pKa = 4.34SLTDD96 pKa = 3.52EE97 pKa = 4.08QLIDD101 pKa = 3.21ILSYY105 pKa = 10.79HH106 pKa = 6.82VIEE109 pKa = 5.59QIIDD113 pKa = 3.78LNQLSAEE120 pKa = 4.18EE121 pKa = 5.03TFTSMQGDD129 pKa = 3.6NIFFVISNDD138 pKa = 3.01SLYY141 pKa = 10.49INGSNYY147 pKa = 10.08VGGVGASNGVLYY159 pKa = 9.01ATDD162 pKa = 3.27QVLFPDD168 pKa = 4.78AYY170 pKa = 11.24LDD172 pKa = 3.75VTGLVHH178 pKa = 7.42KK179 pKa = 10.44RR180 pKa = 11.84KK181 pKa = 9.35QLSEE185 pKa = 4.05LDD187 pKa = 3.55KK188 pKa = 11.54SIDD191 pKa = 3.6NTGLASTLEE200 pKa = 4.48DD201 pKa = 3.05TSSAYY206 pKa = 9.75TIFAPSNTALQNAEE220 pKa = 4.39LSADD224 pKa = 3.5TLKK227 pKa = 10.98YY228 pKa = 10.24HH229 pKa = 5.65VVSGKK234 pKa = 10.21LLSQDD239 pKa = 3.62FSSSQTYY246 pKa = 6.76TTLNGKK252 pKa = 8.02EE253 pKa = 4.06LSVEE257 pKa = 4.11VSGSSVTINGEE268 pKa = 4.06AAVTTADD275 pKa = 3.22IEE277 pKa = 4.61GVNGVVHH284 pKa = 7.33IIDD287 pKa = 3.66TALAPPSEE295 pKa = 4.32
Molecular weight: 31.45 kDa
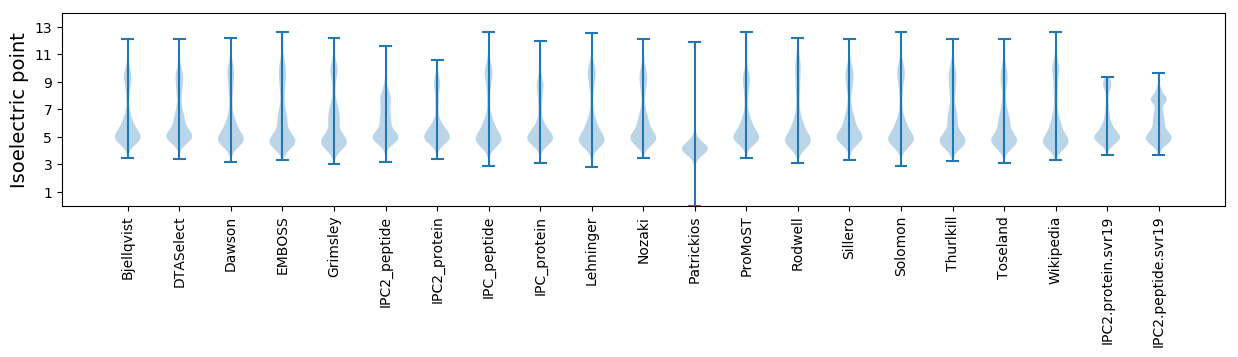
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A521B8R2|A0A521B8R2_9BACT Uncharacterized protein OS=Aliifodinibius sediminis OX=1214077 GN=SAMN06265218_102270 PE=4 SV=1
MM1 pKa = 8.07GDD3 pKa = 3.29QLSNNGIRR11 pKa = 11.84SQGKK15 pKa = 6.87VHH17 pKa = 6.84PLYY20 pKa = 10.87FLGLTSAVAILFVLNLSLGSVSIPFPEE47 pKa = 4.98LLDD50 pKa = 3.26ILMGGEE56 pKa = 4.16AGNTAWEE63 pKa = 4.46KK64 pKa = 10.23IVINIRR70 pKa = 11.84LPRR73 pKa = 11.84AATAVMAGSALAVSGLLMQTLFRR96 pKa = 11.84NPLAGPSVLGITAGASLGVAVVMLAAGSITTVFAIRR132 pKa = 11.84QLSVLGSGVIILAASVGSAVVLLLILLISLKK163 pKa = 10.42VRR165 pKa = 11.84DD166 pKa = 4.9NITLLIIGLMVGNMTIALVSIWQYY190 pKa = 9.99FSRR193 pKa = 11.84PEE195 pKa = 3.85QIQDD199 pKa = 3.36YY200 pKa = 9.66LIWTFGSLGGVSLGQLWVLGVVTVAGGGIAMVLSKK235 pKa = 10.43PLNGLLLGKK244 pKa = 10.22NYY246 pKa = 9.59ARR248 pKa = 11.84SMGIPVSRR256 pKa = 11.84ARR258 pKa = 11.84LWIIISTSLLAGGITAFCGPIGFVGIAVPHH288 pKa = 6.0LTRR291 pKa = 11.84SLLGTNEE298 pKa = 3.79HH299 pKa = 6.57RR300 pKa = 11.84VLIPGTLLMGALLLLACDD318 pKa = 5.51IIAQMPGSTITLPISAVTSMIGSPVVIWVIIRR350 pKa = 11.84RR351 pKa = 11.84KK352 pKa = 8.56NLKK355 pKa = 9.99AAFF358 pKa = 3.96
MM1 pKa = 8.07GDD3 pKa = 3.29QLSNNGIRR11 pKa = 11.84SQGKK15 pKa = 6.87VHH17 pKa = 6.84PLYY20 pKa = 10.87FLGLTSAVAILFVLNLSLGSVSIPFPEE47 pKa = 4.98LLDD50 pKa = 3.26ILMGGEE56 pKa = 4.16AGNTAWEE63 pKa = 4.46KK64 pKa = 10.23IVINIRR70 pKa = 11.84LPRR73 pKa = 11.84AATAVMAGSALAVSGLLMQTLFRR96 pKa = 11.84NPLAGPSVLGITAGASLGVAVVMLAAGSITTVFAIRR132 pKa = 11.84QLSVLGSGVIILAASVGSAVVLLLILLISLKK163 pKa = 10.42VRR165 pKa = 11.84DD166 pKa = 4.9NITLLIIGLMVGNMTIALVSIWQYY190 pKa = 9.99FSRR193 pKa = 11.84PEE195 pKa = 3.85QIQDD199 pKa = 3.36YY200 pKa = 9.66LIWTFGSLGGVSLGQLWVLGVVTVAGGGIAMVLSKK235 pKa = 10.43PLNGLLLGKK244 pKa = 10.22NYY246 pKa = 9.59ARR248 pKa = 11.84SMGIPVSRR256 pKa = 11.84ARR258 pKa = 11.84LWIIISTSLLAGGITAFCGPIGFVGIAVPHH288 pKa = 6.0LTRR291 pKa = 11.84SLLGTNEE298 pKa = 3.79HH299 pKa = 6.57RR300 pKa = 11.84VLIPGTLLMGALLLLACDD318 pKa = 5.51IIAQMPGSTITLPISAVTSMIGSPVVIWVIIRR350 pKa = 11.84RR351 pKa = 11.84KK352 pKa = 8.56NLKK355 pKa = 9.99AAFF358 pKa = 3.96
Molecular weight: 37.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1296162 |
27 |
2887 |
339.1 |
38.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.291 ± 0.042 | 0.674 ± 0.012 |
5.919 ± 0.034 | 7.346 ± 0.046 |
4.377 ± 0.03 | 7.355 ± 0.034 |
2.184 ± 0.021 | 6.565 ± 0.036 |
5.136 ± 0.042 | 9.492 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.471 ± 0.02 | 4.366 ± 0.029 |
4.198 ± 0.024 | 4.068 ± 0.028 |
5.374 ± 0.028 | 6.497 ± 0.029 |
5.33 ± 0.025 | 6.321 ± 0.027 |
1.346 ± 0.015 | 3.694 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
