
Lutibacter oricola
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Lutibacter
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
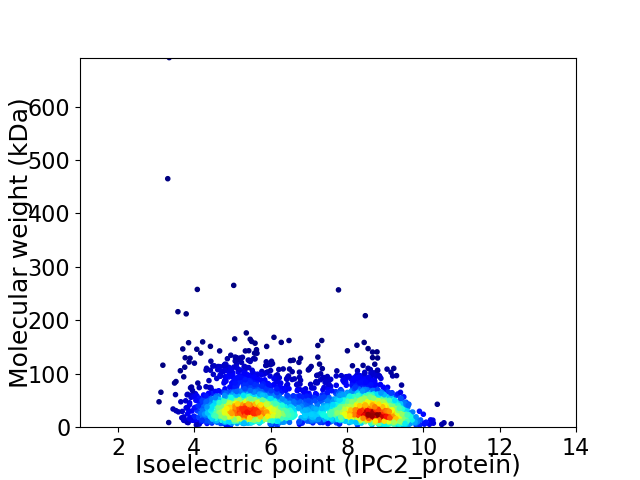
Virtual 2D-PAGE plot for 3464 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3G2N9|A0A1H3G2N9_9FLAO Uncharacterized protein OS=Lutibacter oricola OX=762486 GN=SAMN05444411_11311 PE=4 SV=1
MM1 pKa = 7.21KK2 pKa = 10.09QIKK5 pKa = 9.53YY6 pKa = 9.77LISAIFLFAAVWSCTEE22 pKa = 4.39DD23 pKa = 3.34EE24 pKa = 4.98LGNADD29 pKa = 4.07FVDD32 pKa = 4.66TIVAPTEE39 pKa = 3.78VSAMFNVTQDD49 pKa = 3.05NTGLVTIAPTSQGAVTYY66 pKa = 9.59TVDD69 pKa = 4.72LGDD72 pKa = 4.04GSDD75 pKa = 4.98PITLKK80 pKa = 10.61QGEE83 pKa = 4.53SIKK86 pKa = 11.08NIFAEE91 pKa = 4.36GEE93 pKa = 4.29YY94 pKa = 9.97QVKK97 pKa = 8.32ITAIGVTGLTTSVTLPLVVSFKK119 pKa = 10.96APEE122 pKa = 3.95NLVVEE127 pKa = 4.58ILNDD131 pKa = 3.52EE132 pKa = 4.38AVSKK136 pKa = 10.12QVNVTATADD145 pKa = 3.34NAINFDD151 pKa = 3.71VYY153 pKa = 10.91FGVEE157 pKa = 4.42GVTEE161 pKa = 5.45PITGVIGEE169 pKa = 4.6TVSYY173 pKa = 9.98VYY175 pKa = 9.76PEE177 pKa = 4.0PGIYY181 pKa = 8.99TIKK184 pKa = 10.68VVAKK188 pKa = 9.34GAAIEE193 pKa = 4.1TTEE196 pKa = 4.11YY197 pKa = 10.41IEE199 pKa = 4.13EE200 pKa = 4.37FEE202 pKa = 4.27VTEE205 pKa = 4.19IEE207 pKa = 4.36EE208 pKa = 4.44PLEE211 pKa = 4.26KK212 pKa = 10.72APTPPYY218 pKa = 9.94RR219 pKa = 11.84NEE221 pKa = 3.7VDD223 pKa = 3.72VVSIFSDD230 pKa = 3.6AYY232 pKa = 9.68TNITVNEE239 pKa = 4.18WNPGWGQSTTLTDD252 pKa = 4.11FEE254 pKa = 4.81VDD256 pKa = 2.88GDD258 pKa = 4.38NILKK262 pKa = 10.3YY263 pKa = 10.93DD264 pKa = 3.88FLNYY268 pKa = 9.57TGIVTDD274 pKa = 4.05YY275 pKa = 11.6DD276 pKa = 3.71NPTNLAQMEE285 pKa = 4.6FVHH288 pKa = 6.97FDD290 pKa = 2.94YY291 pKa = 8.2WTNDD295 pKa = 3.26AEE297 pKa = 4.41SLSLKK302 pKa = 10.02IVNTSQADD310 pKa = 3.68GTPEE314 pKa = 4.0KK315 pKa = 10.18EE316 pKa = 4.03SEE318 pKa = 4.14IAVGDD323 pKa = 3.53ITAGEE328 pKa = 4.24WVSVEE333 pKa = 3.96IPLSDD338 pKa = 3.43FTTNMSGVTQFLFVSNGVTVFIDD361 pKa = 2.92NFYY364 pKa = 10.58FYY366 pKa = 10.54KK367 pKa = 10.6QPTTVSEE374 pKa = 4.74GIVGVWKK381 pKa = 9.96LAPIAGALKK390 pKa = 10.55VGPNPNDD397 pKa = 3.61GSWWANSAEE406 pKa = 4.28DD407 pKa = 3.64VTGRR411 pKa = 11.84DD412 pKa = 3.72CLFDD416 pKa = 4.1DD417 pKa = 4.87SYY419 pKa = 12.23VFGADD424 pKa = 3.15GTFKK428 pKa = 11.11NILDD432 pKa = 3.73GSTWVEE438 pKa = 4.16GWQGGSDD445 pKa = 3.49SCATPVAPHH454 pKa = 7.12DD455 pKa = 4.31GSNPATFVYY464 pKa = 10.57DD465 pKa = 3.86EE466 pKa = 4.64AAGKK470 pKa = 8.14VTLNGLGAYY479 pKa = 10.31LGIPKK484 pKa = 10.13AYY486 pKa = 10.23DD487 pKa = 3.39GGEE490 pKa = 4.1LGSPADD496 pKa = 3.88APSAINYY503 pKa = 8.19DD504 pKa = 3.54VTLSNNDD511 pKa = 2.84SAMTVVVNIGSGYY524 pKa = 6.42WTFEE528 pKa = 4.0LVKK531 pKa = 10.44DD532 pKa = 4.14GAPAYY537 pKa = 10.64SPLEE541 pKa = 4.18GTWVVAPEE549 pKa = 4.17AGSLGVGPGQGDD561 pKa = 3.95TSWWTIDD568 pKa = 3.33AAGVTQRR575 pKa = 11.84ACFFDD580 pKa = 3.59DD581 pKa = 3.65TFVFGADD588 pKa = 3.13GSFMNVLGADD598 pKa = 3.29TWIEE602 pKa = 4.03GWQGGTDD609 pKa = 3.19ACGVPVAPHH618 pKa = 7.12DD619 pKa = 4.08GMNPATYY626 pKa = 8.88TYY628 pKa = 11.11DD629 pKa = 3.43EE630 pKa = 4.61AAGKK634 pKa = 8.45LTLNGIGAYY643 pKa = 10.34LGIPKK648 pKa = 10.01AYY650 pKa = 10.5NEE652 pKa = 4.74GEE654 pKa = 4.16LASPADD660 pKa = 3.83APASITYY667 pKa = 10.43DD668 pKa = 3.18VTLANNDD675 pKa = 3.09SEE677 pKa = 4.61MTVDD681 pKa = 3.02IWVGGDD687 pKa = 2.88AWWRR691 pKa = 11.84FKK693 pKa = 9.91MVKK696 pKa = 9.95EE697 pKa = 4.35GTSTGGGSGTGGGTSSGTQIDD718 pKa = 4.35LPVDD722 pKa = 4.38FEE724 pKa = 4.7SATIDD729 pKa = 3.37YY730 pKa = 8.23TLTDD734 pKa = 3.71FGEE737 pKa = 4.35NLSSIVEE744 pKa = 4.3DD745 pKa = 3.57PTNASNTVAKK755 pKa = 10.16VVKK758 pKa = 8.93TAAATTWAGTTIGTDD773 pKa = 2.73TGFATNIPISLTNSIITVRR792 pKa = 11.84VWSPDD797 pKa = 2.49AGAPIRR803 pKa = 11.84LKK805 pKa = 10.58IEE807 pKa = 4.3DD808 pKa = 4.59ANDD811 pKa = 3.39ATHH814 pKa = 5.87TCEE817 pKa = 4.24TEE819 pKa = 4.06VNTTVAGGWEE829 pKa = 4.17TIEE832 pKa = 5.57FDD834 pKa = 4.13FTNQATGTEE843 pKa = 4.23LLSVGLDD850 pKa = 3.27NGWVYY855 pKa = 11.43NKK857 pKa = 10.7ASIFFNFGTDD867 pKa = 3.04GATAGEE873 pKa = 4.0KK874 pKa = 9.59TYY876 pKa = 11.21YY877 pKa = 10.01FDD879 pKa = 6.31DD880 pKa = 4.63VIFINNN886 pKa = 3.32
MM1 pKa = 7.21KK2 pKa = 10.09QIKK5 pKa = 9.53YY6 pKa = 9.77LISAIFLFAAVWSCTEE22 pKa = 4.39DD23 pKa = 3.34EE24 pKa = 4.98LGNADD29 pKa = 4.07FVDD32 pKa = 4.66TIVAPTEE39 pKa = 3.78VSAMFNVTQDD49 pKa = 3.05NTGLVTIAPTSQGAVTYY66 pKa = 9.59TVDD69 pKa = 4.72LGDD72 pKa = 4.04GSDD75 pKa = 4.98PITLKK80 pKa = 10.61QGEE83 pKa = 4.53SIKK86 pKa = 11.08NIFAEE91 pKa = 4.36GEE93 pKa = 4.29YY94 pKa = 9.97QVKK97 pKa = 8.32ITAIGVTGLTTSVTLPLVVSFKK119 pKa = 10.96APEE122 pKa = 3.95NLVVEE127 pKa = 4.58ILNDD131 pKa = 3.52EE132 pKa = 4.38AVSKK136 pKa = 10.12QVNVTATADD145 pKa = 3.34NAINFDD151 pKa = 3.71VYY153 pKa = 10.91FGVEE157 pKa = 4.42GVTEE161 pKa = 5.45PITGVIGEE169 pKa = 4.6TVSYY173 pKa = 9.98VYY175 pKa = 9.76PEE177 pKa = 4.0PGIYY181 pKa = 8.99TIKK184 pKa = 10.68VVAKK188 pKa = 9.34GAAIEE193 pKa = 4.1TTEE196 pKa = 4.11YY197 pKa = 10.41IEE199 pKa = 4.13EE200 pKa = 4.37FEE202 pKa = 4.27VTEE205 pKa = 4.19IEE207 pKa = 4.36EE208 pKa = 4.44PLEE211 pKa = 4.26KK212 pKa = 10.72APTPPYY218 pKa = 9.94RR219 pKa = 11.84NEE221 pKa = 3.7VDD223 pKa = 3.72VVSIFSDD230 pKa = 3.6AYY232 pKa = 9.68TNITVNEE239 pKa = 4.18WNPGWGQSTTLTDD252 pKa = 4.11FEE254 pKa = 4.81VDD256 pKa = 2.88GDD258 pKa = 4.38NILKK262 pKa = 10.3YY263 pKa = 10.93DD264 pKa = 3.88FLNYY268 pKa = 9.57TGIVTDD274 pKa = 4.05YY275 pKa = 11.6DD276 pKa = 3.71NPTNLAQMEE285 pKa = 4.6FVHH288 pKa = 6.97FDD290 pKa = 2.94YY291 pKa = 8.2WTNDD295 pKa = 3.26AEE297 pKa = 4.41SLSLKK302 pKa = 10.02IVNTSQADD310 pKa = 3.68GTPEE314 pKa = 4.0KK315 pKa = 10.18EE316 pKa = 4.03SEE318 pKa = 4.14IAVGDD323 pKa = 3.53ITAGEE328 pKa = 4.24WVSVEE333 pKa = 3.96IPLSDD338 pKa = 3.43FTTNMSGVTQFLFVSNGVTVFIDD361 pKa = 2.92NFYY364 pKa = 10.58FYY366 pKa = 10.54KK367 pKa = 10.6QPTTVSEE374 pKa = 4.74GIVGVWKK381 pKa = 9.96LAPIAGALKK390 pKa = 10.55VGPNPNDD397 pKa = 3.61GSWWANSAEE406 pKa = 4.28DD407 pKa = 3.64VTGRR411 pKa = 11.84DD412 pKa = 3.72CLFDD416 pKa = 4.1DD417 pKa = 4.87SYY419 pKa = 12.23VFGADD424 pKa = 3.15GTFKK428 pKa = 11.11NILDD432 pKa = 3.73GSTWVEE438 pKa = 4.16GWQGGSDD445 pKa = 3.49SCATPVAPHH454 pKa = 7.12DD455 pKa = 4.31GSNPATFVYY464 pKa = 10.57DD465 pKa = 3.86EE466 pKa = 4.64AAGKK470 pKa = 8.14VTLNGLGAYY479 pKa = 10.31LGIPKK484 pKa = 10.13AYY486 pKa = 10.23DD487 pKa = 3.39GGEE490 pKa = 4.1LGSPADD496 pKa = 3.88APSAINYY503 pKa = 8.19DD504 pKa = 3.54VTLSNNDD511 pKa = 2.84SAMTVVVNIGSGYY524 pKa = 6.42WTFEE528 pKa = 4.0LVKK531 pKa = 10.44DD532 pKa = 4.14GAPAYY537 pKa = 10.64SPLEE541 pKa = 4.18GTWVVAPEE549 pKa = 4.17AGSLGVGPGQGDD561 pKa = 3.95TSWWTIDD568 pKa = 3.33AAGVTQRR575 pKa = 11.84ACFFDD580 pKa = 3.59DD581 pKa = 3.65TFVFGADD588 pKa = 3.13GSFMNVLGADD598 pKa = 3.29TWIEE602 pKa = 4.03GWQGGTDD609 pKa = 3.19ACGVPVAPHH618 pKa = 7.12DD619 pKa = 4.08GMNPATYY626 pKa = 8.88TYY628 pKa = 11.11DD629 pKa = 3.43EE630 pKa = 4.61AAGKK634 pKa = 8.45LTLNGIGAYY643 pKa = 10.34LGIPKK648 pKa = 10.01AYY650 pKa = 10.5NEE652 pKa = 4.74GEE654 pKa = 4.16LASPADD660 pKa = 3.83APASITYY667 pKa = 10.43DD668 pKa = 3.18VTLANNDD675 pKa = 3.09SEE677 pKa = 4.61MTVDD681 pKa = 3.02IWVGGDD687 pKa = 2.88AWWRR691 pKa = 11.84FKK693 pKa = 9.91MVKK696 pKa = 9.95EE697 pKa = 4.35GTSTGGGSGTGGGTSSGTQIDD718 pKa = 4.35LPVDD722 pKa = 4.38FEE724 pKa = 4.7SATIDD729 pKa = 3.37YY730 pKa = 8.23TLTDD734 pKa = 3.71FGEE737 pKa = 4.35NLSSIVEE744 pKa = 4.3DD745 pKa = 3.57PTNASNTVAKK755 pKa = 10.16VVKK758 pKa = 8.93TAAATTWAGTTIGTDD773 pKa = 2.73TGFATNIPISLTNSIITVRR792 pKa = 11.84VWSPDD797 pKa = 2.49AGAPIRR803 pKa = 11.84LKK805 pKa = 10.58IEE807 pKa = 4.3DD808 pKa = 4.59ANDD811 pKa = 3.39ATHH814 pKa = 5.87TCEE817 pKa = 4.24TEE819 pKa = 4.06VNTTVAGGWEE829 pKa = 4.17TIEE832 pKa = 5.57FDD834 pKa = 4.13FTNQATGTEE843 pKa = 4.23LLSVGLDD850 pKa = 3.27NGWVYY855 pKa = 11.43NKK857 pKa = 10.7ASIFFNFGTDD867 pKa = 3.04GATAGEE873 pKa = 4.0KK874 pKa = 9.59TYY876 pKa = 11.21YY877 pKa = 10.01FDD879 pKa = 6.31DD880 pKa = 4.63VIFINNN886 pKa = 3.32
Molecular weight: 94.28 kDa
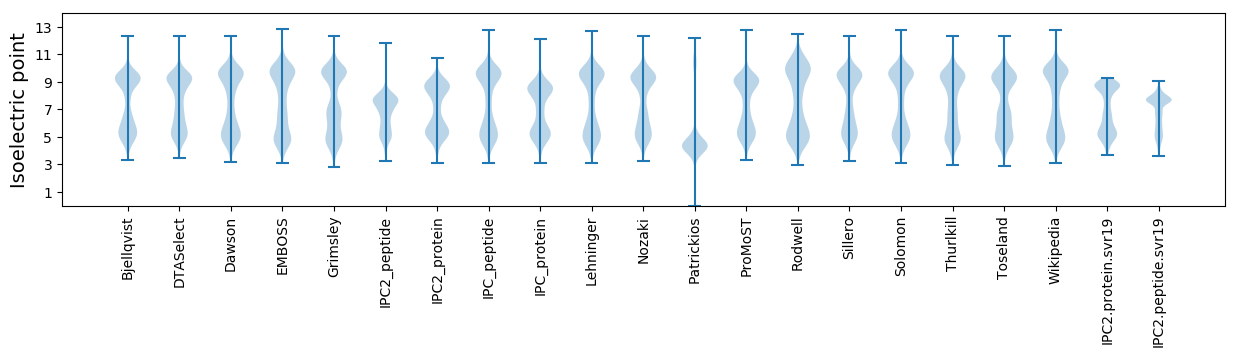
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H2Z930|A0A1H2Z930_9FLAO Nitroreductase OS=Lutibacter oricola OX=762486 GN=SAMN05444411_103179 PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.39KK31 pKa = 10.55SLLAPKK37 pKa = 9.91KK38 pKa = 10.38RR39 pKa = 11.84SGGRR43 pKa = 11.84NNKK46 pKa = 9.29GKK48 pKa = 8.29MTMRR52 pKa = 11.84YY53 pKa = 9.26IGGGHH58 pKa = 6.45KK59 pKa = 9.63KK60 pKa = 9.44HH61 pKa = 6.1YY62 pKa = 10.08RR63 pKa = 11.84IIDD66 pKa = 3.83FKK68 pKa = 10.83RR69 pKa = 11.84NKK71 pKa = 9.58HH72 pKa = 5.09GVPATVKK79 pKa = 10.58SIEE82 pKa = 4.0YY83 pKa = 10.15DD84 pKa = 3.43PNRR87 pKa = 11.84TAFIALLNYY96 pKa = 10.38ADD98 pKa = 3.65GEE100 pKa = 4.2KK101 pKa = 10.21RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLKK111 pKa = 10.38AGQTVISGEE120 pKa = 4.13GATPDD125 pKa = 3.47VGNTMLLSEE134 pKa = 4.99IPLGTTISCIEE145 pKa = 3.88LHH147 pKa = 6.73PGQGAVIARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84EE168 pKa = 4.45GKK170 pKa = 10.09YY171 pKa = 9.05ATIKK175 pKa = 10.52LPSGEE180 pKa = 4.19TRR182 pKa = 11.84LILLTCSATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 9.87AGRR210 pKa = 11.84TRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVVMNPVDD229 pKa = 3.47HH230 pKa = 7.05PMGGGEE236 pKa = 4.28GKK238 pKa = 10.51SSGGHH243 pKa = 4.59PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.41GFKK256 pKa = 9.24TRR258 pKa = 11.84SKK260 pKa = 9.37TKK262 pKa = 10.4ASNKK266 pKa = 9.95YY267 pKa = 9.56IIEE270 pKa = 4.06RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.39KK31 pKa = 10.55SLLAPKK37 pKa = 9.91KK38 pKa = 10.38RR39 pKa = 11.84SGGRR43 pKa = 11.84NNKK46 pKa = 9.29GKK48 pKa = 8.29MTMRR52 pKa = 11.84YY53 pKa = 9.26IGGGHH58 pKa = 6.45KK59 pKa = 9.63KK60 pKa = 9.44HH61 pKa = 6.1YY62 pKa = 10.08RR63 pKa = 11.84IIDD66 pKa = 3.83FKK68 pKa = 10.83RR69 pKa = 11.84NKK71 pKa = 9.58HH72 pKa = 5.09GVPATVKK79 pKa = 10.58SIEE82 pKa = 4.0YY83 pKa = 10.15DD84 pKa = 3.43PNRR87 pKa = 11.84TAFIALLNYY96 pKa = 10.38ADD98 pKa = 3.65GEE100 pKa = 4.2KK101 pKa = 10.21RR102 pKa = 11.84YY103 pKa = 10.74VIAQNGLKK111 pKa = 10.38AGQTVISGEE120 pKa = 4.13GATPDD125 pKa = 3.47VGNTMLLSEE134 pKa = 4.99IPLGTTISCIEE145 pKa = 3.88LHH147 pKa = 6.73PGQGAVIARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84EE168 pKa = 4.45GKK170 pKa = 10.09YY171 pKa = 9.05ATIKK175 pKa = 10.52LPSGEE180 pKa = 4.19TRR182 pKa = 11.84LILLTCSATIGVVSNSDD199 pKa = 3.21HH200 pKa = 6.09QLLVSGKK207 pKa = 9.87AGRR210 pKa = 11.84TRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVVMNPVDD229 pKa = 3.47HH230 pKa = 7.05PMGGGEE236 pKa = 4.28GKK238 pKa = 10.51SSGGHH243 pKa = 4.59PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.41GFKK256 pKa = 9.24TRR258 pKa = 11.84SKK260 pKa = 9.37TKK262 pKa = 10.4ASNKK266 pKa = 9.95YY267 pKa = 9.56IIEE270 pKa = 4.06RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
Molecular weight: 29.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1260383 |
25 |
6593 |
363.9 |
41.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.891 ± 0.042 | 0.772 ± 0.014 |
5.279 ± 0.037 | 6.73 ± 0.043 |
5.208 ± 0.034 | 6.35 ± 0.038 |
1.657 ± 0.019 | 8.206 ± 0.039 |
8.719 ± 0.069 | 8.936 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.068 ± 0.022 | 6.929 ± 0.046 |
3.25 ± 0.026 | 2.974 ± 0.021 |
2.979 ± 0.024 | 6.61 ± 0.036 |
5.852 ± 0.059 | 6.324 ± 0.04 |
1.132 ± 0.016 | 4.134 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
