
Camel associated porprismacovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 7.91
Get precalculated fractions of proteins
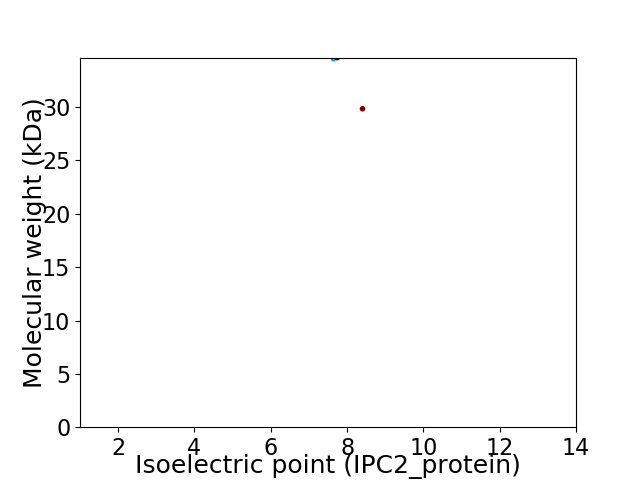
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
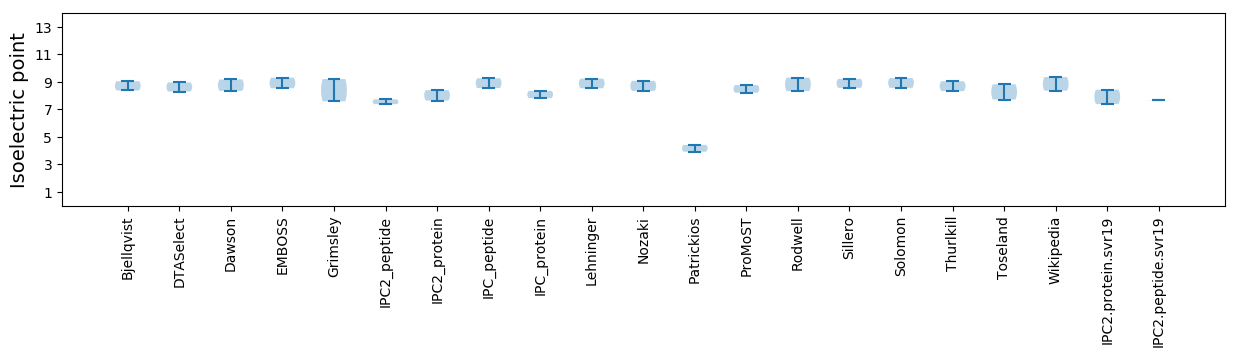
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1ENU8|A0A0A1ENU8_9VIRU Putative replicase protein OS=Camel associated porprismacovirus 2 OX=2170106 GN=rep PE=4 SV=1
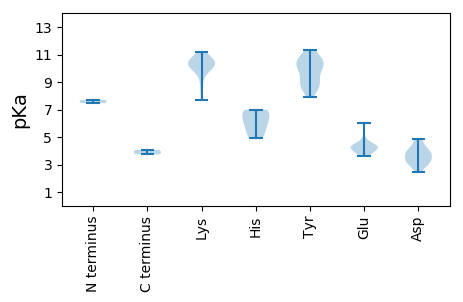
MM1 pKa = 7.48KK2 pKa = 10.43FKK4 pKa = 10.43FQSFFDD10 pKa = 3.98VATSASSMQVIQWSVGGQEE29 pKa = 3.79VLEE32 pKa = 4.3RR33 pKa = 11.84CRR35 pKa = 11.84HH36 pKa = 4.92LFGAYY41 pKa = 9.38KK42 pKa = 10.11YY43 pKa = 10.17FKK45 pKa = 10.22LGKK48 pKa = 9.43ISVKK52 pKa = 10.1FVPASTLPVDD62 pKa = 4.11PQGLSYY68 pKa = 11.14DD69 pKa = 3.73ADD71 pKa = 4.07DD72 pKa = 4.89PQTVDD77 pKa = 4.88PRR79 pKa = 11.84DD80 pKa = 3.3QMNPGLVRR88 pKa = 11.84ITNGEE93 pKa = 4.33DD94 pKa = 2.66IFTNVSAISAANQEE108 pKa = 4.26KK109 pKa = 10.73VYY111 pKa = 10.82NAMMLDD117 pKa = 3.81PRR119 pKa = 11.84WSKK122 pKa = 11.16FMLQSGFRR130 pKa = 11.84RR131 pKa = 11.84EE132 pKa = 4.19AYY134 pKa = 9.1PLYY137 pKa = 9.39WQIGQLSQDD146 pKa = 3.69YY147 pKa = 9.21WPGATVNTPVVGEE160 pKa = 4.23AQATATRR167 pKa = 11.84PLYY170 pKa = 10.05IRR172 pKa = 11.84NVNGAFDD179 pKa = 3.73TAYY182 pKa = 8.31GTSGDD187 pKa = 3.53SSARR191 pKa = 11.84GIFQTGHH198 pKa = 6.08RR199 pKa = 11.84GKK201 pKa = 10.8LGFLPTDD208 pKa = 4.13YY209 pKa = 11.31YY210 pKa = 10.07QTTVNTRR217 pKa = 11.84GTASANYY224 pKa = 8.62GANSVPTVSVITCVLPKK241 pKa = 10.18AYY243 pKa = 8.45KK244 pKa = 7.7TVYY247 pKa = 8.71YY248 pKa = 10.19YY249 pKa = 10.71RR250 pKa = 11.84VFVTEE255 pKa = 3.83EE256 pKa = 4.0VIFAGLRR263 pKa = 11.84SSPMAEE269 pKa = 3.85ANGSQTYY276 pKa = 10.79GLDD279 pKa = 3.04QFLFANMINIFPGTASTRR297 pKa = 11.84PSSIEE302 pKa = 3.62PNKK305 pKa = 10.53QNDD308 pKa = 4.17GANLPP313 pKa = 4.06
MM1 pKa = 7.48KK2 pKa = 10.43FKK4 pKa = 10.43FQSFFDD10 pKa = 3.98VATSASSMQVIQWSVGGQEE29 pKa = 3.79VLEE32 pKa = 4.3RR33 pKa = 11.84CRR35 pKa = 11.84HH36 pKa = 4.92LFGAYY41 pKa = 9.38KK42 pKa = 10.11YY43 pKa = 10.17FKK45 pKa = 10.22LGKK48 pKa = 9.43ISVKK52 pKa = 10.1FVPASTLPVDD62 pKa = 4.11PQGLSYY68 pKa = 11.14DD69 pKa = 3.73ADD71 pKa = 4.07DD72 pKa = 4.89PQTVDD77 pKa = 4.88PRR79 pKa = 11.84DD80 pKa = 3.3QMNPGLVRR88 pKa = 11.84ITNGEE93 pKa = 4.33DD94 pKa = 2.66IFTNVSAISAANQEE108 pKa = 4.26KK109 pKa = 10.73VYY111 pKa = 10.82NAMMLDD117 pKa = 3.81PRR119 pKa = 11.84WSKK122 pKa = 11.16FMLQSGFRR130 pKa = 11.84RR131 pKa = 11.84EE132 pKa = 4.19AYY134 pKa = 9.1PLYY137 pKa = 9.39WQIGQLSQDD146 pKa = 3.69YY147 pKa = 9.21WPGATVNTPVVGEE160 pKa = 4.23AQATATRR167 pKa = 11.84PLYY170 pKa = 10.05IRR172 pKa = 11.84NVNGAFDD179 pKa = 3.73TAYY182 pKa = 8.31GTSGDD187 pKa = 3.53SSARR191 pKa = 11.84GIFQTGHH198 pKa = 6.08RR199 pKa = 11.84GKK201 pKa = 10.8LGFLPTDD208 pKa = 4.13YY209 pKa = 11.31YY210 pKa = 10.07QTTVNTRR217 pKa = 11.84GTASANYY224 pKa = 8.62GANSVPTVSVITCVLPKK241 pKa = 10.18AYY243 pKa = 8.45KK244 pKa = 7.7TVYY247 pKa = 8.71YY248 pKa = 10.19YY249 pKa = 10.71RR250 pKa = 11.84VFVTEE255 pKa = 3.83EE256 pKa = 4.0VIFAGLRR263 pKa = 11.84SSPMAEE269 pKa = 3.85ANGSQTYY276 pKa = 10.79GLDD279 pKa = 3.04QFLFANMINIFPGTASTRR297 pKa = 11.84PSSIEE302 pKa = 3.62PNKK305 pKa = 10.53QNDD308 pKa = 4.17GANLPP313 pKa = 4.06
Molecular weight: 34.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1ENU8|A0A0A1ENU8_9VIRU Putative replicase protein OS=Camel associated porprismacovirus 2 OX=2170106 GN=rep PE=4 SV=1
MM1 pKa = 7.71GIYY4 pKa = 9.28MVTAPRR10 pKa = 11.84KK11 pKa = 8.81GSKK14 pKa = 10.23LGTGKK19 pKa = 10.49EE20 pKa = 4.02FMKK23 pKa = 10.15WRR25 pKa = 11.84TFFYY29 pKa = 10.62INDD32 pKa = 3.41VHH34 pKa = 6.96KK35 pKa = 8.42WTLGAEE41 pKa = 4.24TGKK44 pKa = 10.75GGYY47 pKa = 8.53KK48 pKa = 9.55HH49 pKa = 5.4WQVRR53 pKa = 11.84LQISGSDD60 pKa = 3.08NEE62 pKa = 4.51VFNKK66 pKa = 10.16IKK68 pKa = 10.32DD69 pKa = 3.7EE70 pKa = 4.21NMFPTAHH77 pKa = 6.65IEE79 pKa = 4.16KK80 pKa = 10.76GSDD83 pKa = 2.47TWSYY87 pKa = 8.8EE88 pKa = 4.51CKK90 pKa = 10.57EE91 pKa = 4.02KK92 pKa = 9.96MHH94 pKa = 6.16WTSNDD99 pKa = 2.96TPNILRR105 pKa = 11.84VRR107 pKa = 11.84FGHH110 pKa = 5.28MRR112 pKa = 11.84WYY114 pKa = 10.05QKK116 pKa = 11.09DD117 pKa = 3.37VIEE120 pKa = 4.48AVRR123 pKa = 11.84RR124 pKa = 11.84QNDD127 pKa = 3.2RR128 pKa = 11.84QIAYY132 pKa = 8.88WYY134 pKa = 10.07DD135 pKa = 3.19PEE137 pKa = 4.47GNKK140 pKa = 10.04GKK142 pKa = 10.15SWLVNHH148 pKa = 6.98LFEE151 pKa = 6.01SCQAWYY157 pKa = 10.48VPPTLKK163 pKa = 10.54SVEE166 pKa = 5.0GIIQWVASVYY176 pKa = 10.55INNNYY181 pKa = 10.09RR182 pKa = 11.84EE183 pKa = 4.25ILVIDD188 pKa = 4.65IPRR191 pKa = 11.84SWKK194 pKa = 9.63WSTEE198 pKa = 3.66LYY200 pKa = 9.73TAIEE204 pKa = 4.35TIKK207 pKa = 10.95DD208 pKa = 3.31GLVYY212 pKa = 10.41DD213 pKa = 4.43PRR215 pKa = 11.84YY216 pKa = 7.95HH217 pKa = 6.9AQMINIRR224 pKa = 11.84GVKK227 pKa = 9.71ILVLCNHH234 pKa = 6.06EE235 pKa = 4.6PKK237 pKa = 10.53LDD239 pKa = 3.83ALSADD244 pKa = 2.93RR245 pKa = 11.84WYY247 pKa = 10.94AVAPALTT254 pKa = 3.74
MM1 pKa = 7.71GIYY4 pKa = 9.28MVTAPRR10 pKa = 11.84KK11 pKa = 8.81GSKK14 pKa = 10.23LGTGKK19 pKa = 10.49EE20 pKa = 4.02FMKK23 pKa = 10.15WRR25 pKa = 11.84TFFYY29 pKa = 10.62INDD32 pKa = 3.41VHH34 pKa = 6.96KK35 pKa = 8.42WTLGAEE41 pKa = 4.24TGKK44 pKa = 10.75GGYY47 pKa = 8.53KK48 pKa = 9.55HH49 pKa = 5.4WQVRR53 pKa = 11.84LQISGSDD60 pKa = 3.08NEE62 pKa = 4.51VFNKK66 pKa = 10.16IKK68 pKa = 10.32DD69 pKa = 3.7EE70 pKa = 4.21NMFPTAHH77 pKa = 6.65IEE79 pKa = 4.16KK80 pKa = 10.76GSDD83 pKa = 2.47TWSYY87 pKa = 8.8EE88 pKa = 4.51CKK90 pKa = 10.57EE91 pKa = 4.02KK92 pKa = 9.96MHH94 pKa = 6.16WTSNDD99 pKa = 2.96TPNILRR105 pKa = 11.84VRR107 pKa = 11.84FGHH110 pKa = 5.28MRR112 pKa = 11.84WYY114 pKa = 10.05QKK116 pKa = 11.09DD117 pKa = 3.37VIEE120 pKa = 4.48AVRR123 pKa = 11.84RR124 pKa = 11.84QNDD127 pKa = 3.2RR128 pKa = 11.84QIAYY132 pKa = 8.88WYY134 pKa = 10.07DD135 pKa = 3.19PEE137 pKa = 4.47GNKK140 pKa = 10.04GKK142 pKa = 10.15SWLVNHH148 pKa = 6.98LFEE151 pKa = 6.01SCQAWYY157 pKa = 10.48VPPTLKK163 pKa = 10.54SVEE166 pKa = 5.0GIIQWVASVYY176 pKa = 10.55INNNYY181 pKa = 10.09RR182 pKa = 11.84EE183 pKa = 4.25ILVIDD188 pKa = 4.65IPRR191 pKa = 11.84SWKK194 pKa = 9.63WSTEE198 pKa = 3.66LYY200 pKa = 9.73TAIEE204 pKa = 4.35TIKK207 pKa = 10.95DD208 pKa = 3.31GLVYY212 pKa = 10.41DD213 pKa = 4.43PRR215 pKa = 11.84YY216 pKa = 7.95HH217 pKa = 6.9AQMINIRR224 pKa = 11.84GVKK227 pKa = 9.71ILVLCNHH234 pKa = 6.06EE235 pKa = 4.6PKK237 pKa = 10.53LDD239 pKa = 3.83ALSADD244 pKa = 2.93RR245 pKa = 11.84WYY247 pKa = 10.94AVAPALTT254 pKa = 3.74
Molecular weight: 29.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
567 |
254 |
313 |
283.5 |
32.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.231 ± 1.105 | 0.882 ± 0.192 |
4.938 ± 0.116 | 4.409 ± 0.961 |
4.409 ± 1.062 | 7.231 ± 0.599 |
1.764 ± 0.89 | 5.644 ± 1.18 |
5.644 ± 1.433 | 5.82 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.646 ± 0.071 | 5.467 ± 0.029 |
5.115 ± 0.757 | 4.586 ± 0.923 |
5.115 ± 0.255 | 6.702 ± 1.018 |
6.702 ± 0.765 | 7.231 ± 0.346 |
2.998 ± 1.362 | 5.467 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
