
Fowlpox virus (strain NVSL) (FPV)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Pokkesviricetes; Chitovirales; Poxviridae; Chordopoxvirinae; Avipoxvirus; Fowlpox virus
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
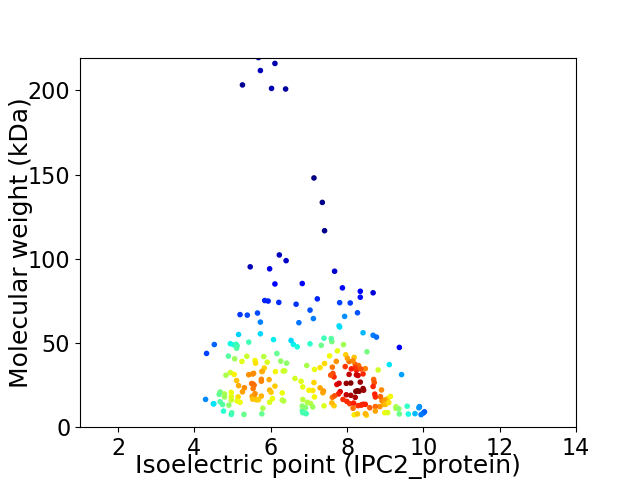
Virtual 2D-PAGE plot for 251 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
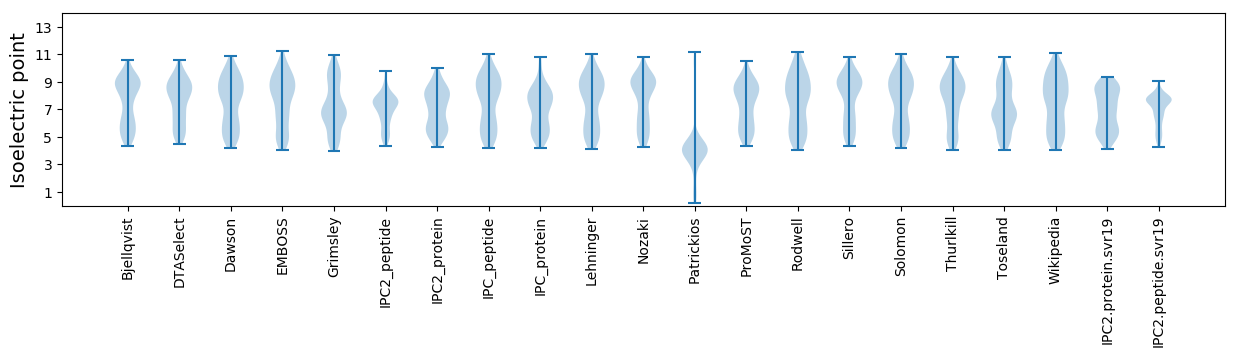
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9J572|Q9J572_FOWPN ORF FPV159 N1R/p28 gene family protein OS=Fowlpox virus (strain NVSL) OX=928301 GN=FPV159 PE=4 SV=1
MM1 pKa = 7.4IRR3 pKa = 11.84IIILSLLFINVTTDD17 pKa = 3.0SQEE20 pKa = 3.88SSKK23 pKa = 11.12NIQNVLHH30 pKa = 4.88VTEE33 pKa = 4.43YY34 pKa = 11.14SRR36 pKa = 11.84TGVTACSLHH45 pKa = 6.74CFDD48 pKa = 5.25RR49 pKa = 11.84SKK51 pKa = 11.54GLDD54 pKa = 3.29QPKK57 pKa = 8.87TFILPGKK64 pKa = 9.66YY65 pKa = 9.99SNNSIKK71 pKa = 10.87LEE73 pKa = 4.12VAIDD77 pKa = 3.92TYY79 pKa = 11.58KK80 pKa = 10.6KK81 pKa = 10.7DD82 pKa = 3.28SDD84 pKa = 3.66FSYY87 pKa = 10.74SHH89 pKa = 7.57PCQIFQFCVSGNFSGKK105 pKa = 10.17RR106 pKa = 11.84FDD108 pKa = 3.75HH109 pKa = 6.1YY110 pKa = 11.22LYY112 pKa = 10.49GYY114 pKa = 7.55TISGFIDD121 pKa = 3.63IAPKK125 pKa = 10.21YY126 pKa = 8.37YY127 pKa = 10.66SGMSISTITVMPLQEE142 pKa = 4.73GSLKK146 pKa = 10.54HH147 pKa = 7.11DD148 pKa = 4.52DD149 pKa = 4.33ADD151 pKa = 5.16DD152 pKa = 3.68YY153 pKa = 11.7DD154 pKa = 5.13YY155 pKa = 11.81DD156 pKa = 4.79DD157 pKa = 5.44DD158 pKa = 4.15CVPYY162 pKa = 10.82KK163 pKa = 9.49EE164 pKa = 4.43TQPRR168 pKa = 11.84HH169 pKa = 4.94MPEE172 pKa = 3.82SVIKK176 pKa = 9.7EE177 pKa = 4.11GCKK180 pKa = 9.99PIPLPRR186 pKa = 11.84YY187 pKa = 9.74DD188 pKa = 4.68EE189 pKa = 5.08NDD191 pKa = 3.57DD192 pKa = 4.01PTCIMYY198 pKa = 9.47WDD200 pKa = 4.67HH201 pKa = 7.43SWDD204 pKa = 3.52NYY206 pKa = 9.9CNVGFFNSLQSDD218 pKa = 4.07HH219 pKa = 6.78NPLVFPLTSYY229 pKa = 11.6SDD231 pKa = 3.37INNAFHH237 pKa = 7.41AFQSSYY243 pKa = 10.74CRR245 pKa = 11.84SLGFNQSYY253 pKa = 9.06SVCVSIGDD261 pKa = 3.77TPFEE265 pKa = 4.35VTYY268 pKa = 10.58HH269 pKa = 6.23SYY271 pKa = 11.43EE272 pKa = 4.34SVTVDD277 pKa = 4.52QLLQEE282 pKa = 4.31IKK284 pKa = 9.71TLYY287 pKa = 11.06GEE289 pKa = 4.31DD290 pKa = 3.48AVYY293 pKa = 10.79GLPFRR298 pKa = 11.84NITIMARR305 pKa = 11.84TRR307 pKa = 11.84IQSLPLTNNTCIPKK321 pKa = 9.81QDD323 pKa = 4.4DD324 pKa = 3.71ADD326 pKa = 4.33DD327 pKa = 4.28VDD329 pKa = 5.3DD330 pKa = 6.47ADD332 pKa = 5.75DD333 pKa = 5.13VDD335 pKa = 5.16DD336 pKa = 6.63ADD338 pKa = 6.03DD339 pKa = 5.77ADD341 pKa = 5.77DD342 pKa = 5.39DD343 pKa = 5.13DD344 pKa = 6.79DD345 pKa = 5.74YY346 pKa = 11.91EE347 pKa = 5.48LYY349 pKa = 10.97VEE351 pKa = 4.42TTPRR355 pKa = 11.84VPTARR360 pKa = 11.84KK361 pKa = 9.53KK362 pKa = 10.48PVTEE366 pKa = 4.25EE367 pKa = 3.94YY368 pKa = 11.27NDD370 pKa = 3.43IFSSFDD376 pKa = 3.26NFDD379 pKa = 3.59MKK381 pKa = 10.82KK382 pKa = 10.03KK383 pKa = 10.59
MM1 pKa = 7.4IRR3 pKa = 11.84IIILSLLFINVTTDD17 pKa = 3.0SQEE20 pKa = 3.88SSKK23 pKa = 11.12NIQNVLHH30 pKa = 4.88VTEE33 pKa = 4.43YY34 pKa = 11.14SRR36 pKa = 11.84TGVTACSLHH45 pKa = 6.74CFDD48 pKa = 5.25RR49 pKa = 11.84SKK51 pKa = 11.54GLDD54 pKa = 3.29QPKK57 pKa = 8.87TFILPGKK64 pKa = 9.66YY65 pKa = 9.99SNNSIKK71 pKa = 10.87LEE73 pKa = 4.12VAIDD77 pKa = 3.92TYY79 pKa = 11.58KK80 pKa = 10.6KK81 pKa = 10.7DD82 pKa = 3.28SDD84 pKa = 3.66FSYY87 pKa = 10.74SHH89 pKa = 7.57PCQIFQFCVSGNFSGKK105 pKa = 10.17RR106 pKa = 11.84FDD108 pKa = 3.75HH109 pKa = 6.1YY110 pKa = 11.22LYY112 pKa = 10.49GYY114 pKa = 7.55TISGFIDD121 pKa = 3.63IAPKK125 pKa = 10.21YY126 pKa = 8.37YY127 pKa = 10.66SGMSISTITVMPLQEE142 pKa = 4.73GSLKK146 pKa = 10.54HH147 pKa = 7.11DD148 pKa = 4.52DD149 pKa = 4.33ADD151 pKa = 5.16DD152 pKa = 3.68YY153 pKa = 11.7DD154 pKa = 5.13YY155 pKa = 11.81DD156 pKa = 4.79DD157 pKa = 5.44DD158 pKa = 4.15CVPYY162 pKa = 10.82KK163 pKa = 9.49EE164 pKa = 4.43TQPRR168 pKa = 11.84HH169 pKa = 4.94MPEE172 pKa = 3.82SVIKK176 pKa = 9.7EE177 pKa = 4.11GCKK180 pKa = 9.99PIPLPRR186 pKa = 11.84YY187 pKa = 9.74DD188 pKa = 4.68EE189 pKa = 5.08NDD191 pKa = 3.57DD192 pKa = 4.01PTCIMYY198 pKa = 9.47WDD200 pKa = 4.67HH201 pKa = 7.43SWDD204 pKa = 3.52NYY206 pKa = 9.9CNVGFFNSLQSDD218 pKa = 4.07HH219 pKa = 6.78NPLVFPLTSYY229 pKa = 11.6SDD231 pKa = 3.37INNAFHH237 pKa = 7.41AFQSSYY243 pKa = 10.74CRR245 pKa = 11.84SLGFNQSYY253 pKa = 9.06SVCVSIGDD261 pKa = 3.77TPFEE265 pKa = 4.35VTYY268 pKa = 10.58HH269 pKa = 6.23SYY271 pKa = 11.43EE272 pKa = 4.34SVTVDD277 pKa = 4.52QLLQEE282 pKa = 4.31IKK284 pKa = 9.71TLYY287 pKa = 11.06GEE289 pKa = 4.31DD290 pKa = 3.48AVYY293 pKa = 10.79GLPFRR298 pKa = 11.84NITIMARR305 pKa = 11.84TRR307 pKa = 11.84IQSLPLTNNTCIPKK321 pKa = 9.81QDD323 pKa = 4.4DD324 pKa = 3.71ADD326 pKa = 4.33DD327 pKa = 4.28VDD329 pKa = 5.3DD330 pKa = 6.47ADD332 pKa = 5.75DD333 pKa = 5.13VDD335 pKa = 5.16DD336 pKa = 6.63ADD338 pKa = 6.03DD339 pKa = 5.77ADD341 pKa = 5.77DD342 pKa = 5.39DD343 pKa = 5.13DD344 pKa = 6.79DD345 pKa = 5.74YY346 pKa = 11.91EE347 pKa = 5.48LYY349 pKa = 10.97VEE351 pKa = 4.42TTPRR355 pKa = 11.84VPTARR360 pKa = 11.84KK361 pKa = 9.53KK362 pKa = 10.48PVTEE366 pKa = 4.25EE367 pKa = 3.94YY368 pKa = 11.27NDD370 pKa = 3.43IFSSFDD376 pKa = 3.26NFDD379 pKa = 3.59MKK381 pKa = 10.82KK382 pKa = 10.03KK383 pKa = 10.59
Molecular weight: 43.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9ICF5|Q9ICF5_FOWPN ORF FPV005 EFc gene family protein OS=Fowlpox virus (strain NVSL) OX=928301 GN=FPV005 PE=4 SV=1
MM1 pKa = 7.31NNDD4 pKa = 3.74TIFTLFYY11 pKa = 10.24CKK13 pKa = 10.01NKK15 pKa = 10.26KK16 pKa = 8.03YY17 pKa = 10.88VRR19 pKa = 11.84GEE21 pKa = 3.65GGRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84GKK28 pKa = 8.62TGILLFHH35 pKa = 7.38PINHH39 pKa = 6.04RR40 pKa = 11.84VIGTSAHH47 pKa = 4.55QCYY50 pKa = 7.68KK51 pKa = 8.7TRR53 pKa = 11.84RR54 pKa = 11.84IGFKK58 pKa = 10.18LYY60 pKa = 10.64AVAPRR65 pKa = 11.84HH66 pKa = 5.64VSTIRR71 pKa = 11.84CGRR74 pKa = 11.84SHH76 pKa = 6.53SAHH79 pKa = 6.32RR80 pKa = 11.84VDD82 pKa = 4.51KK83 pKa = 11.1FSFSFQKK90 pKa = 10.53VDD92 pKa = 4.42FHH94 pKa = 7.29CIAGSGAA101 pKa = 3.35
MM1 pKa = 7.31NNDD4 pKa = 3.74TIFTLFYY11 pKa = 10.24CKK13 pKa = 10.01NKK15 pKa = 10.26KK16 pKa = 8.03YY17 pKa = 10.88VRR19 pKa = 11.84GEE21 pKa = 3.65GGRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84GKK28 pKa = 8.62TGILLFHH35 pKa = 7.38PINHH39 pKa = 6.04RR40 pKa = 11.84VIGTSAHH47 pKa = 4.55QCYY50 pKa = 7.68KK51 pKa = 8.7TRR53 pKa = 11.84RR54 pKa = 11.84IGFKK58 pKa = 10.18LYY60 pKa = 10.64AVAPRR65 pKa = 11.84HH66 pKa = 5.64VSTIRR71 pKa = 11.84CGRR74 pKa = 11.84SHH76 pKa = 6.53SAHH79 pKa = 6.32RR80 pKa = 11.84VDD82 pKa = 4.51KK83 pKa = 11.1FSFSFQKK90 pKa = 10.53VDD92 pKa = 4.42FHH94 pKa = 7.29CIAGSGAA101 pKa = 3.35
Molecular weight: 11.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
80735 |
60 |
1949 |
321.7 |
37.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.433 ± 0.097 | 2.2 ± 0.067 |
6.4 ± 0.107 | 5.599 ± 0.104 |
4.255 ± 0.118 | 4.003 ± 0.11 |
2.019 ± 0.064 | 10.035 ± 0.187 |
8.224 ± 0.125 | 9.109 ± 0.173 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.555 ± 0.06 | 7.235 ± 0.148 |
3.151 ± 0.093 | 1.99 ± 0.069 |
4.132 ± 0.093 | 7.889 ± 0.164 |
5.724 ± 0.121 | 5.672 ± 0.097 |
0.666 ± 0.038 | 5.708 ± 0.096 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
