
Rubripirellula amarantea
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Rubripirellula
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
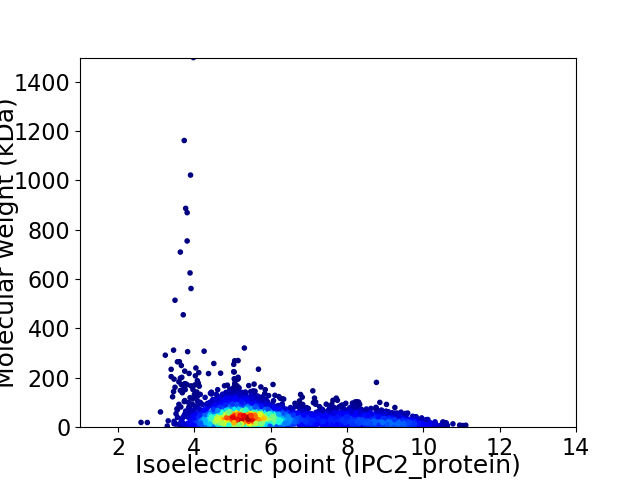
Virtual 2D-PAGE plot for 5179 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C5WU88|A0A5C5WU88_9BACT NAD_binding_9 domain-containing protein OS=Rubripirellula amarantea OX=2527999 GN=Pla22_14220 PE=4 SV=1
MM1 pKa = 7.03NRR3 pKa = 11.84SYY5 pKa = 11.17LCKK8 pKa = 10.1RR9 pKa = 11.84SQGFFGLWSIAIALAAMPPDD29 pKa = 3.43QSHH32 pKa = 6.29AQTIVATNPPTQITTEE48 pKa = 4.0TFYY51 pKa = 10.93EE52 pKa = 3.75IDD54 pKa = 3.42VAYY57 pKa = 10.69SIADD61 pKa = 3.56DD62 pKa = 5.76AIVQVQLFQMLEE74 pKa = 4.02NSWAFIDD81 pKa = 4.4QVWEE85 pKa = 4.02PVLDD89 pKa = 4.16GSTSIAIPFYY99 pKa = 10.61VPEE102 pKa = 4.1EE103 pKa = 3.96AAPEE107 pKa = 4.14DD108 pKa = 3.73NYY110 pKa = 11.06LWQVILYY117 pKa = 9.9NGSDD121 pKa = 3.34WEE123 pKa = 4.36KK124 pKa = 10.31LDD126 pKa = 4.38EE127 pKa = 4.84EE128 pKa = 4.25IFYY131 pKa = 9.85PITVSANAPDD141 pKa = 4.43VITGSNAPPEE151 pKa = 4.34VQTNEE156 pKa = 4.28SYY158 pKa = 11.33DD159 pKa = 3.38VTVEE163 pKa = 3.93YY164 pKa = 10.18EE165 pKa = 3.77ISVDD169 pKa = 4.68GIVQVQLLDD178 pKa = 3.89SSWNVIDD185 pKa = 4.91QDD187 pKa = 3.57WTSVDD192 pKa = 3.68SGSEE196 pKa = 4.13SATVTIDD203 pKa = 3.53VPIDD207 pKa = 3.28QPLADD212 pKa = 4.22DD213 pKa = 5.16SIWQIMLYY221 pKa = 10.2RR222 pKa = 11.84QDD224 pKa = 3.7PNTWTKK230 pKa = 10.68LDD232 pKa = 4.12EE233 pKa = 5.27IIAEE237 pKa = 4.96GIAIVGNMGGPNLEE251 pKa = 4.12WLPPGNSWALEE262 pKa = 3.99WQDD265 pKa = 3.47EE266 pKa = 4.55FEE268 pKa = 4.52GTGLPEE274 pKa = 3.11NWYY277 pKa = 9.0PHH279 pKa = 6.93LGTNPEE285 pKa = 4.12EE286 pKa = 4.29VSNEE290 pKa = 3.38IAAAAAEE297 pKa = 4.08NRR299 pKa = 11.84IPEE302 pKa = 4.07YY303 pKa = 9.44PLRR306 pKa = 11.84YY307 pKa = 9.68GPYY310 pKa = 9.62DD311 pKa = 3.65GNNAWMFSTGYY322 pKa = 10.03GNHH325 pKa = 6.74WLDD328 pKa = 4.02GSGNLQLQIRR338 pKa = 11.84ADD340 pKa = 3.92LNVDD344 pKa = 4.12LAHH347 pKa = 6.31GKK349 pKa = 9.65KK350 pKa = 9.92VEE352 pKa = 4.07SAYY355 pKa = 11.11LMSGQPVAWDD365 pKa = 3.62DD366 pKa = 3.99SVPGTGVRR374 pKa = 11.84WEE376 pKa = 4.29GTLVSPGDD384 pKa = 3.59GEE386 pKa = 4.76IYY388 pKa = 10.15ISTRR392 pKa = 11.84VKK394 pKa = 8.14TNEE397 pKa = 3.63MVGYY401 pKa = 7.57STWFAFWLFTQTQAYY416 pKa = 9.92DD417 pKa = 3.51RR418 pKa = 11.84VPATGTEE425 pKa = 3.55IDD427 pKa = 4.34IIEE430 pKa = 4.25IPIGEE435 pKa = 4.62PAWMDD440 pKa = 3.28GVFNVAHH447 pKa = 6.52HH448 pKa = 6.61WGVPTQAEE456 pKa = 4.4LNAGIKK462 pKa = 10.35FSEE465 pKa = 4.19SLQYY469 pKa = 10.88SGVIAEE475 pKa = 4.52SLADD479 pKa = 3.65VTADD483 pKa = 3.38EE484 pKa = 4.36YY485 pKa = 10.67HH486 pKa = 6.4TYY488 pKa = 9.96GLRR491 pKa = 11.84WSADD495 pKa = 2.91SMTTYY500 pKa = 11.08VNGVEE505 pKa = 4.42TFTFPDD511 pKa = 3.94NPPGNPVPAIPANPSDD527 pKa = 3.94MMMMLTLEE535 pKa = 4.28FEE537 pKa = 3.86KK538 pKa = 10.98DD539 pKa = 2.78AWVDD543 pKa = 3.37KK544 pKa = 10.73TLDD547 pKa = 3.59DD548 pKa = 3.87QGDD551 pKa = 3.63GRR553 pKa = 11.84FQGTPLEE560 pKa = 4.14EE561 pKa = 4.28DD562 pKa = 2.79ANYY565 pKa = 10.82RR566 pKa = 11.84VMSRR570 pKa = 11.84AQVDD574 pKa = 3.66YY575 pKa = 10.93VRR577 pKa = 11.84IWRR580 pKa = 11.84AQQ582 pKa = 2.99
MM1 pKa = 7.03NRR3 pKa = 11.84SYY5 pKa = 11.17LCKK8 pKa = 10.1RR9 pKa = 11.84SQGFFGLWSIAIALAAMPPDD29 pKa = 3.43QSHH32 pKa = 6.29AQTIVATNPPTQITTEE48 pKa = 4.0TFYY51 pKa = 10.93EE52 pKa = 3.75IDD54 pKa = 3.42VAYY57 pKa = 10.69SIADD61 pKa = 3.56DD62 pKa = 5.76AIVQVQLFQMLEE74 pKa = 4.02NSWAFIDD81 pKa = 4.4QVWEE85 pKa = 4.02PVLDD89 pKa = 4.16GSTSIAIPFYY99 pKa = 10.61VPEE102 pKa = 4.1EE103 pKa = 3.96AAPEE107 pKa = 4.14DD108 pKa = 3.73NYY110 pKa = 11.06LWQVILYY117 pKa = 9.9NGSDD121 pKa = 3.34WEE123 pKa = 4.36KK124 pKa = 10.31LDD126 pKa = 4.38EE127 pKa = 4.84EE128 pKa = 4.25IFYY131 pKa = 9.85PITVSANAPDD141 pKa = 4.43VITGSNAPPEE151 pKa = 4.34VQTNEE156 pKa = 4.28SYY158 pKa = 11.33DD159 pKa = 3.38VTVEE163 pKa = 3.93YY164 pKa = 10.18EE165 pKa = 3.77ISVDD169 pKa = 4.68GIVQVQLLDD178 pKa = 3.89SSWNVIDD185 pKa = 4.91QDD187 pKa = 3.57WTSVDD192 pKa = 3.68SGSEE196 pKa = 4.13SATVTIDD203 pKa = 3.53VPIDD207 pKa = 3.28QPLADD212 pKa = 4.22DD213 pKa = 5.16SIWQIMLYY221 pKa = 10.2RR222 pKa = 11.84QDD224 pKa = 3.7PNTWTKK230 pKa = 10.68LDD232 pKa = 4.12EE233 pKa = 5.27IIAEE237 pKa = 4.96GIAIVGNMGGPNLEE251 pKa = 4.12WLPPGNSWALEE262 pKa = 3.99WQDD265 pKa = 3.47EE266 pKa = 4.55FEE268 pKa = 4.52GTGLPEE274 pKa = 3.11NWYY277 pKa = 9.0PHH279 pKa = 6.93LGTNPEE285 pKa = 4.12EE286 pKa = 4.29VSNEE290 pKa = 3.38IAAAAAEE297 pKa = 4.08NRR299 pKa = 11.84IPEE302 pKa = 4.07YY303 pKa = 9.44PLRR306 pKa = 11.84YY307 pKa = 9.68GPYY310 pKa = 9.62DD311 pKa = 3.65GNNAWMFSTGYY322 pKa = 10.03GNHH325 pKa = 6.74WLDD328 pKa = 4.02GSGNLQLQIRR338 pKa = 11.84ADD340 pKa = 3.92LNVDD344 pKa = 4.12LAHH347 pKa = 6.31GKK349 pKa = 9.65KK350 pKa = 9.92VEE352 pKa = 4.07SAYY355 pKa = 11.11LMSGQPVAWDD365 pKa = 3.62DD366 pKa = 3.99SVPGTGVRR374 pKa = 11.84WEE376 pKa = 4.29GTLVSPGDD384 pKa = 3.59GEE386 pKa = 4.76IYY388 pKa = 10.15ISTRR392 pKa = 11.84VKK394 pKa = 8.14TNEE397 pKa = 3.63MVGYY401 pKa = 7.57STWFAFWLFTQTQAYY416 pKa = 9.92DD417 pKa = 3.51RR418 pKa = 11.84VPATGTEE425 pKa = 3.55IDD427 pKa = 4.34IIEE430 pKa = 4.25IPIGEE435 pKa = 4.62PAWMDD440 pKa = 3.28GVFNVAHH447 pKa = 6.52HH448 pKa = 6.61WGVPTQAEE456 pKa = 4.4LNAGIKK462 pKa = 10.35FSEE465 pKa = 4.19SLQYY469 pKa = 10.88SGVIAEE475 pKa = 4.52SLADD479 pKa = 3.65VTADD483 pKa = 3.38EE484 pKa = 4.36YY485 pKa = 10.67HH486 pKa = 6.4TYY488 pKa = 9.96GLRR491 pKa = 11.84WSADD495 pKa = 2.91SMTTYY500 pKa = 11.08VNGVEE505 pKa = 4.42TFTFPDD511 pKa = 3.94NPPGNPVPAIPANPSDD527 pKa = 3.94MMMMLTLEE535 pKa = 4.28FEE537 pKa = 3.86KK538 pKa = 10.98DD539 pKa = 2.78AWVDD543 pKa = 3.37KK544 pKa = 10.73TLDD547 pKa = 3.59DD548 pKa = 3.87QGDD551 pKa = 3.63GRR553 pKa = 11.84FQGTPLEE560 pKa = 4.14EE561 pKa = 4.28DD562 pKa = 2.79ANYY565 pKa = 10.82RR566 pKa = 11.84VMSRR570 pKa = 11.84AQVDD574 pKa = 3.66YY575 pKa = 10.93VRR577 pKa = 11.84IWRR580 pKa = 11.84AQQ582 pKa = 2.99
Molecular weight: 64.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C5WII3|A0A5C5WII3_9BACT PEP-CTERM motif protein OS=Rubripirellula amarantea OX=2527999 GN=Pla22_48190 PE=4 SV=1
MM1 pKa = 6.89TLAVAMTFAVPMKK14 pKa = 8.98FTVPMKK20 pKa = 9.32FTVPMTFTVPMTFTVPGPRR39 pKa = 11.84NQRR42 pKa = 11.84AQPRR46 pKa = 11.84GRR48 pKa = 11.84RR49 pKa = 11.84SKK51 pKa = 9.64PARR54 pKa = 11.84PVVTTARR61 pKa = 11.84AAGAA65 pKa = 3.57
MM1 pKa = 6.89TLAVAMTFAVPMKK14 pKa = 8.98FTVPMKK20 pKa = 9.32FTVPMTFTVPMTFTVPGPRR39 pKa = 11.84NQRR42 pKa = 11.84AQPRR46 pKa = 11.84GRR48 pKa = 11.84RR49 pKa = 11.84SKK51 pKa = 9.64PARR54 pKa = 11.84PVVTTARR61 pKa = 11.84AAGAA65 pKa = 3.57
Molecular weight: 7.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2058698 |
29 |
14289 |
397.5 |
43.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.376 ± 0.037 | 1.054 ± 0.019 |
6.661 ± 0.056 | 5.925 ± 0.032 |
3.727 ± 0.019 | 7.722 ± 0.063 |
2.132 ± 0.027 | 5.349 ± 0.022 |
3.534 ± 0.037 | 9.37 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.327 ± 0.028 | 3.461 ± 0.033 |
4.877 ± 0.031 | 3.893 ± 0.024 |
6.247 ± 0.051 | 7.283 ± 0.036 |
5.862 ± 0.057 | 7.44 ± 0.029 |
1.39 ± 0.017 | 2.368 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
