
Chimeric virus 14
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
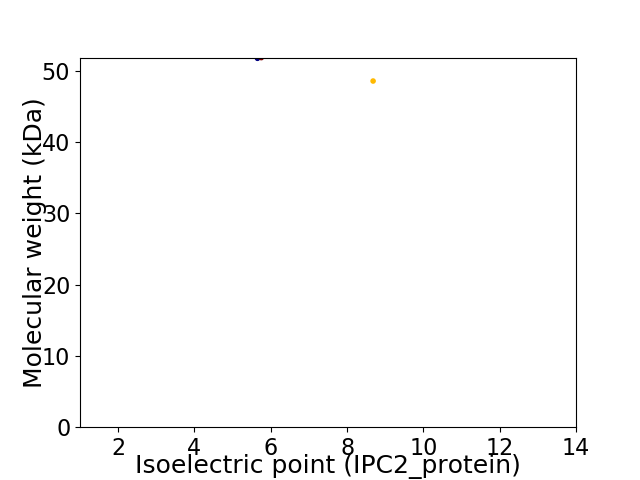
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5ALV7|A0A0C5ALV7_9VIRU Putative replication protein OS=Chimeric virus 14 OX=1608440 GN=rep PE=3 SV=1
MM1 pKa = 7.44SAPWDD6 pKa = 3.84NDD8 pKa = 3.2SEE10 pKa = 4.51EE11 pKa = 4.17YY12 pKa = 11.02SNEE15 pKa = 3.83EE16 pKa = 4.33KK17 pKa = 9.97IDD19 pKa = 3.48RR20 pKa = 11.84APNRR24 pKa = 11.84KK25 pKa = 8.95VKK27 pKa = 10.42IKK29 pKa = 10.3IAEE32 pKa = 4.21DD33 pKa = 4.0PYY35 pKa = 11.24EE36 pKa = 4.39EE37 pKa = 4.59PSEE40 pKa = 4.52DD41 pKa = 5.67GIIQDD46 pKa = 3.88LDD48 pKa = 3.74EE49 pKa = 6.32KK50 pKa = 11.31EE51 pKa = 4.5EE52 pKa = 3.98EE53 pKa = 4.3LDD55 pKa = 3.79NFSKK59 pKa = 10.86SALRR63 pKa = 11.84SNGVFKK69 pKa = 10.9INNRR73 pKa = 11.84QVMLTYY79 pKa = 8.94KK80 pKa = 9.2THH82 pKa = 7.23LDD84 pKa = 3.46KK85 pKa = 11.17EE86 pKa = 4.72DD87 pKa = 3.32YY88 pKa = 10.6RR89 pKa = 11.84SFVDD93 pKa = 3.74EE94 pKa = 4.67LAPFDD99 pKa = 4.2EE100 pKa = 5.56LFIAHH105 pKa = 6.77EE106 pKa = 4.24NADD109 pKa = 3.61LKK111 pKa = 11.28NPYY114 pKa = 9.39KK115 pKa = 9.71HH116 pKa = 4.63THH118 pKa = 6.09AYY120 pKa = 9.13IKK122 pKa = 9.22FTIPFVSSDD131 pKa = 3.1SRR133 pKa = 11.84VFDD136 pKa = 3.44WDD138 pKa = 3.3VSEE141 pKa = 4.31TQNIHH146 pKa = 4.9PHH148 pKa = 5.93ISVLGAKK155 pKa = 9.83KK156 pKa = 10.23VDD158 pKa = 4.24EE159 pKa = 4.79IKK161 pKa = 10.07MKK163 pKa = 10.84YY164 pKa = 9.94YY165 pKa = 9.95ISKK168 pKa = 9.96EE169 pKa = 4.09DD170 pKa = 3.71KK171 pKa = 11.19SPDD174 pKa = 3.28LAMLRR179 pKa = 11.84VSCDD183 pKa = 2.87DD184 pKa = 3.33WNKK187 pKa = 10.85ANGNKK192 pKa = 9.45ARR194 pKa = 11.84LGKK197 pKa = 10.51GLADD201 pKa = 3.91DD202 pKa = 3.76QVSLLEE208 pKa = 4.25RR209 pKa = 11.84VCSYY213 pKa = 9.39KK214 pKa = 10.34TYY216 pKa = 11.01QEE218 pKa = 4.11MLKK221 pKa = 10.08HH222 pKa = 5.55LCKK225 pKa = 10.57EE226 pKa = 4.24PGHH229 pKa = 5.61ATGLRR234 pKa = 11.84EE235 pKa = 3.9LWEE238 pKa = 4.41SKK240 pKa = 10.76LIAEE244 pKa = 4.36KK245 pKa = 10.59PKK247 pKa = 10.56IKK249 pKa = 10.29FSEE252 pKa = 4.09IAEE255 pKa = 4.3FTWQKK260 pKa = 8.73TMRR263 pKa = 11.84DD264 pKa = 3.4KK265 pKa = 11.4LLAPEE270 pKa = 4.09WNHH273 pKa = 7.38RR274 pKa = 11.84ILTYY278 pKa = 10.39IVDD281 pKa = 3.87KK282 pKa = 11.07KK283 pKa = 10.95GGSGKK288 pKa = 8.56STFADD293 pKa = 3.41YY294 pKa = 10.05MVNTYY299 pKa = 10.23NALYY303 pKa = 9.66LSSVTSSRR311 pKa = 11.84DD312 pKa = 2.88IATLIDD318 pKa = 3.44EE319 pKa = 4.53HH320 pKa = 6.76LKK322 pKa = 11.06CGGSAKK328 pKa = 10.57YY329 pKa = 10.11IILDD333 pKa = 3.76LPRR336 pKa = 11.84ASKK339 pKa = 9.51AHH341 pKa = 6.07KK342 pKa = 9.51MWDD345 pKa = 3.4TVEE348 pKa = 4.63ALLNGKK354 pKa = 8.24LTVGKK359 pKa = 9.35WKK361 pKa = 10.55GKK363 pKa = 8.75PLHH366 pKa = 6.37FDD368 pKa = 3.27KK369 pKa = 10.95PRR371 pKa = 11.84LIVFANFAPPLPGDD385 pKa = 3.66EE386 pKa = 4.34SRR388 pKa = 11.84AADD391 pKa = 3.63VCEE394 pKa = 4.51DD395 pKa = 3.76NCVLSPDD402 pKa = 3.9RR403 pKa = 11.84WDD405 pKa = 3.16IWDD408 pKa = 4.06IVCLDD413 pKa = 3.46AAKK416 pKa = 10.69AHH418 pKa = 6.68DD419 pKa = 4.45GEE421 pKa = 4.74HH422 pKa = 6.5DD423 pKa = 4.04RR424 pKa = 11.84IMVNRR429 pKa = 11.84ANPHH433 pKa = 6.79RR434 pKa = 11.84DD435 pKa = 3.69LNWVKK440 pKa = 10.47PKK442 pKa = 10.53RR443 pKa = 11.84FQNLDD448 pKa = 3.44YY449 pKa = 11.43
MM1 pKa = 7.44SAPWDD6 pKa = 3.84NDD8 pKa = 3.2SEE10 pKa = 4.51EE11 pKa = 4.17YY12 pKa = 11.02SNEE15 pKa = 3.83EE16 pKa = 4.33KK17 pKa = 9.97IDD19 pKa = 3.48RR20 pKa = 11.84APNRR24 pKa = 11.84KK25 pKa = 8.95VKK27 pKa = 10.42IKK29 pKa = 10.3IAEE32 pKa = 4.21DD33 pKa = 4.0PYY35 pKa = 11.24EE36 pKa = 4.39EE37 pKa = 4.59PSEE40 pKa = 4.52DD41 pKa = 5.67GIIQDD46 pKa = 3.88LDD48 pKa = 3.74EE49 pKa = 6.32KK50 pKa = 11.31EE51 pKa = 4.5EE52 pKa = 3.98EE53 pKa = 4.3LDD55 pKa = 3.79NFSKK59 pKa = 10.86SALRR63 pKa = 11.84SNGVFKK69 pKa = 10.9INNRR73 pKa = 11.84QVMLTYY79 pKa = 8.94KK80 pKa = 9.2THH82 pKa = 7.23LDD84 pKa = 3.46KK85 pKa = 11.17EE86 pKa = 4.72DD87 pKa = 3.32YY88 pKa = 10.6RR89 pKa = 11.84SFVDD93 pKa = 3.74EE94 pKa = 4.67LAPFDD99 pKa = 4.2EE100 pKa = 5.56LFIAHH105 pKa = 6.77EE106 pKa = 4.24NADD109 pKa = 3.61LKK111 pKa = 11.28NPYY114 pKa = 9.39KK115 pKa = 9.71HH116 pKa = 4.63THH118 pKa = 6.09AYY120 pKa = 9.13IKK122 pKa = 9.22FTIPFVSSDD131 pKa = 3.1SRR133 pKa = 11.84VFDD136 pKa = 3.44WDD138 pKa = 3.3VSEE141 pKa = 4.31TQNIHH146 pKa = 4.9PHH148 pKa = 5.93ISVLGAKK155 pKa = 9.83KK156 pKa = 10.23VDD158 pKa = 4.24EE159 pKa = 4.79IKK161 pKa = 10.07MKK163 pKa = 10.84YY164 pKa = 9.94YY165 pKa = 9.95ISKK168 pKa = 9.96EE169 pKa = 4.09DD170 pKa = 3.71KK171 pKa = 11.19SPDD174 pKa = 3.28LAMLRR179 pKa = 11.84VSCDD183 pKa = 2.87DD184 pKa = 3.33WNKK187 pKa = 10.85ANGNKK192 pKa = 9.45ARR194 pKa = 11.84LGKK197 pKa = 10.51GLADD201 pKa = 3.91DD202 pKa = 3.76QVSLLEE208 pKa = 4.25RR209 pKa = 11.84VCSYY213 pKa = 9.39KK214 pKa = 10.34TYY216 pKa = 11.01QEE218 pKa = 4.11MLKK221 pKa = 10.08HH222 pKa = 5.55LCKK225 pKa = 10.57EE226 pKa = 4.24PGHH229 pKa = 5.61ATGLRR234 pKa = 11.84EE235 pKa = 3.9LWEE238 pKa = 4.41SKK240 pKa = 10.76LIAEE244 pKa = 4.36KK245 pKa = 10.59PKK247 pKa = 10.56IKK249 pKa = 10.29FSEE252 pKa = 4.09IAEE255 pKa = 4.3FTWQKK260 pKa = 8.73TMRR263 pKa = 11.84DD264 pKa = 3.4KK265 pKa = 11.4LLAPEE270 pKa = 4.09WNHH273 pKa = 7.38RR274 pKa = 11.84ILTYY278 pKa = 10.39IVDD281 pKa = 3.87KK282 pKa = 11.07KK283 pKa = 10.95GGSGKK288 pKa = 8.56STFADD293 pKa = 3.41YY294 pKa = 10.05MVNTYY299 pKa = 10.23NALYY303 pKa = 9.66LSSVTSSRR311 pKa = 11.84DD312 pKa = 2.88IATLIDD318 pKa = 3.44EE319 pKa = 4.53HH320 pKa = 6.76LKK322 pKa = 11.06CGGSAKK328 pKa = 10.57YY329 pKa = 10.11IILDD333 pKa = 3.76LPRR336 pKa = 11.84ASKK339 pKa = 9.51AHH341 pKa = 6.07KK342 pKa = 9.51MWDD345 pKa = 3.4TVEE348 pKa = 4.63ALLNGKK354 pKa = 8.24LTVGKK359 pKa = 9.35WKK361 pKa = 10.55GKK363 pKa = 8.75PLHH366 pKa = 6.37FDD368 pKa = 3.27KK369 pKa = 10.95PRR371 pKa = 11.84LIVFANFAPPLPGDD385 pKa = 3.66EE386 pKa = 4.34SRR388 pKa = 11.84AADD391 pKa = 3.63VCEE394 pKa = 4.51DD395 pKa = 3.76NCVLSPDD402 pKa = 3.9RR403 pKa = 11.84WDD405 pKa = 3.16IWDD408 pKa = 4.06IVCLDD413 pKa = 3.46AAKK416 pKa = 10.69AHH418 pKa = 6.68DD419 pKa = 4.45GEE421 pKa = 4.74HH422 pKa = 6.5DD423 pKa = 4.04RR424 pKa = 11.84IMVNRR429 pKa = 11.84ANPHH433 pKa = 6.79RR434 pKa = 11.84DD435 pKa = 3.69LNWVKK440 pKa = 10.47PKK442 pKa = 10.53RR443 pKa = 11.84FQNLDD448 pKa = 3.44YY449 pKa = 11.43
Molecular weight: 51.71 kDa
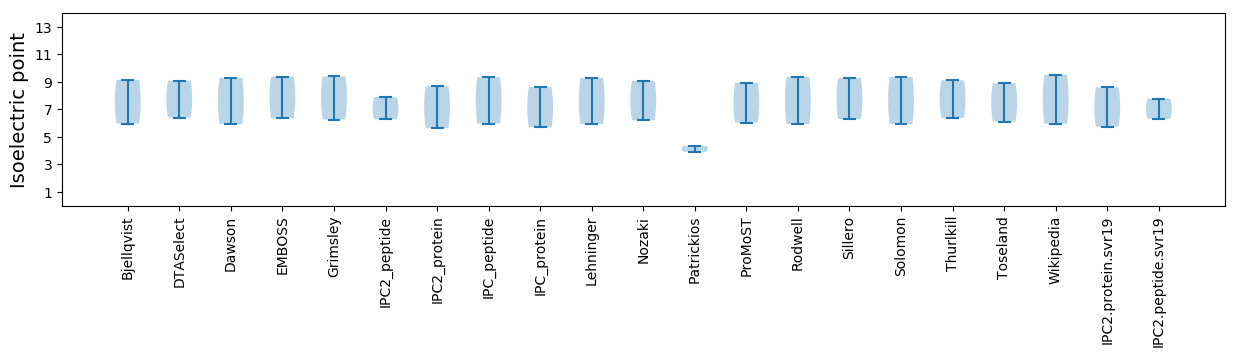
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5ALV7|A0A0C5ALV7_9VIRU Putative replication protein OS=Chimeric virus 14 OX=1608440 GN=rep PE=3 SV=1
MM1 pKa = 6.64STRR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 8.75VKK8 pKa = 10.11KK9 pKa = 10.7SKK11 pKa = 10.86APAPKK16 pKa = 8.99IVYY19 pKa = 9.06VAAPPVAKK27 pKa = 10.0RR28 pKa = 11.84SYY30 pKa = 8.18TRR32 pKa = 11.84KK33 pKa = 10.01SSTAGGLRR41 pKa = 11.84GHH43 pKa = 7.33GDD45 pKa = 3.31YY46 pKa = 10.62TYY48 pKa = 10.86DD49 pKa = 5.25KK50 pKa = 9.69PGPWGNFGRR59 pKa = 11.84SVGKK63 pKa = 9.61QAGAMLGSNYY73 pKa = 10.55GLGKK77 pKa = 10.52AGGAIGSKK85 pKa = 10.28LGGYY89 pKa = 8.35LHH91 pKa = 6.98YY92 pKa = 10.02IGKK95 pKa = 9.43IFGSGDD101 pKa = 3.66YY102 pKa = 8.85VTSSSQVRR110 pKa = 11.84NNILVNQSQVPQFQDD125 pKa = 2.48GKK127 pKa = 7.84NTVRR131 pKa = 11.84IAHH134 pKa = 6.67RR135 pKa = 11.84EE136 pKa = 3.83YY137 pKa = 10.83LGDD140 pKa = 4.72IITSSTPGAFSIEE153 pKa = 4.42SYY155 pKa = 10.09PINPGVSGSFPWLANVVGASFQQYY179 pKa = 9.58RR180 pKa = 11.84INGMVFEE187 pKa = 4.57FRR189 pKa = 11.84SMSADD194 pKa = 2.99ALTSTNTALGTVAMATDD211 pKa = 4.31YY212 pKa = 11.27DD213 pKa = 4.2SKK215 pKa = 11.26DD216 pKa = 3.16TAFTSKK222 pKa = 10.01QQMEE226 pKa = 4.15NTEE229 pKa = 4.36FGVSCKK235 pKa = 9.95PSSCMIHH242 pKa = 7.37GIEE245 pKa = 4.52CARR248 pKa = 11.84SQTSVSEE255 pKa = 4.25LYY257 pKa = 10.5VRR259 pKa = 11.84AFAVPSGADD268 pKa = 3.19PRR270 pKa = 11.84LYY272 pKa = 11.32DD273 pKa = 3.27MGNFYY278 pKa = 10.54IATQGMQGASVNVGEE293 pKa = 4.84LWVSYY298 pKa = 10.68DD299 pKa = 2.9ITFFKK304 pKa = 10.74AIEE307 pKa = 4.01QVPGFIMPLANYY319 pKa = 8.39TPVTASGTAPLGVTRR334 pKa = 11.84GISQPRR340 pKa = 11.84GVDD343 pKa = 3.41QIGLTFTEE351 pKa = 3.81NRR353 pKa = 11.84IIFPYY358 pKa = 9.98NIPIGSTYY366 pKa = 9.6QWTYY370 pKa = 10.03TISTASSALTLPVVTYY386 pKa = 11.08VNGFTDD392 pKa = 4.24LATYY396 pKa = 8.87KK397 pKa = 10.54APSNGEE403 pKa = 3.96VATKK407 pKa = 10.75GLISGMMRR415 pKa = 11.84LTSPGTPSTPPYY427 pKa = 10.9VNLADD432 pKa = 3.84FTVGAISFIALDD444 pKa = 3.64VAQISGFPAEE454 pKa = 4.25PP455 pKa = 3.51
MM1 pKa = 6.64STRR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 8.75VKK8 pKa = 10.11KK9 pKa = 10.7SKK11 pKa = 10.86APAPKK16 pKa = 8.99IVYY19 pKa = 9.06VAAPPVAKK27 pKa = 10.0RR28 pKa = 11.84SYY30 pKa = 8.18TRR32 pKa = 11.84KK33 pKa = 10.01SSTAGGLRR41 pKa = 11.84GHH43 pKa = 7.33GDD45 pKa = 3.31YY46 pKa = 10.62TYY48 pKa = 10.86DD49 pKa = 5.25KK50 pKa = 9.69PGPWGNFGRR59 pKa = 11.84SVGKK63 pKa = 9.61QAGAMLGSNYY73 pKa = 10.55GLGKK77 pKa = 10.52AGGAIGSKK85 pKa = 10.28LGGYY89 pKa = 8.35LHH91 pKa = 6.98YY92 pKa = 10.02IGKK95 pKa = 9.43IFGSGDD101 pKa = 3.66YY102 pKa = 8.85VTSSSQVRR110 pKa = 11.84NNILVNQSQVPQFQDD125 pKa = 2.48GKK127 pKa = 7.84NTVRR131 pKa = 11.84IAHH134 pKa = 6.67RR135 pKa = 11.84EE136 pKa = 3.83YY137 pKa = 10.83LGDD140 pKa = 4.72IITSSTPGAFSIEE153 pKa = 4.42SYY155 pKa = 10.09PINPGVSGSFPWLANVVGASFQQYY179 pKa = 9.58RR180 pKa = 11.84INGMVFEE187 pKa = 4.57FRR189 pKa = 11.84SMSADD194 pKa = 2.99ALTSTNTALGTVAMATDD211 pKa = 4.31YY212 pKa = 11.27DD213 pKa = 4.2SKK215 pKa = 11.26DD216 pKa = 3.16TAFTSKK222 pKa = 10.01QQMEE226 pKa = 4.15NTEE229 pKa = 4.36FGVSCKK235 pKa = 9.95PSSCMIHH242 pKa = 7.37GIEE245 pKa = 4.52CARR248 pKa = 11.84SQTSVSEE255 pKa = 4.25LYY257 pKa = 10.5VRR259 pKa = 11.84AFAVPSGADD268 pKa = 3.19PRR270 pKa = 11.84LYY272 pKa = 11.32DD273 pKa = 3.27MGNFYY278 pKa = 10.54IATQGMQGASVNVGEE293 pKa = 4.84LWVSYY298 pKa = 10.68DD299 pKa = 2.9ITFFKK304 pKa = 10.74AIEE307 pKa = 4.01QVPGFIMPLANYY319 pKa = 8.39TPVTASGTAPLGVTRR334 pKa = 11.84GISQPRR340 pKa = 11.84GVDD343 pKa = 3.41QIGLTFTEE351 pKa = 3.81NRR353 pKa = 11.84IIFPYY358 pKa = 9.98NIPIGSTYY366 pKa = 9.6QWTYY370 pKa = 10.03TISTASSALTLPVVTYY386 pKa = 11.08VNGFTDD392 pKa = 4.24LATYY396 pKa = 8.87KK397 pKa = 10.54APSNGEE403 pKa = 3.96VATKK407 pKa = 10.75GLISGMMRR415 pKa = 11.84LTSPGTPSTPPYY427 pKa = 10.9VNLADD432 pKa = 3.84FTVGAISFIALDD444 pKa = 3.64VAQISGFPAEE454 pKa = 4.25PP455 pKa = 3.51
Molecular weight: 48.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
904 |
449 |
455 |
452.0 |
50.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.965 ± 0.57 | 1.106 ± 0.308 |
6.416 ± 2.0 | 4.978 ± 1.614 |
3.982 ± 0.437 | 7.412 ± 2.316 |
2.102 ± 0.843 | 5.973 ± 0.124 |
6.969 ± 2.078 | 6.858 ± 1.244 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.323 ± 0.217 | 4.646 ± 0.324 |
5.531 ± 0.581 | 2.765 ± 0.821 |
4.314 ± 0.247 | 8.407 ± 1.174 |
5.973 ± 1.64 | 6.195 ± 0.73 |
1.659 ± 0.538 | 4.425 ± 0.586 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
