
Agrobacterium radiobacter (strain K84 / ATCC BAA-868)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Agrobacterium; Agrobacterium tumefaciens complex; Agrobacterium tumefaciens
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
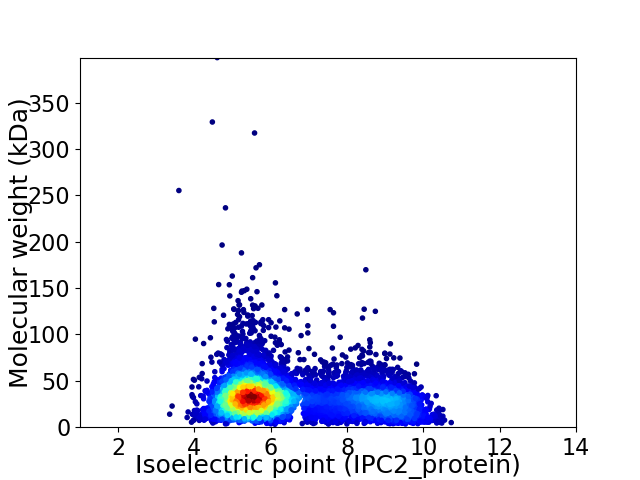
Virtual 2D-PAGE plot for 6674 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B9J9H3|B9J9H3_AGRRK Cell division coordinator CpoB OS=Agrobacterium radiobacter (strain K84 / ATCC BAA-868) OX=311403 GN=ygbF PE=3 SV=1
MM1 pKa = 7.61SIFGSMKK8 pKa = 9.46TAISGMNAQANRR20 pKa = 11.84LATVSDD26 pKa = 4.49NIANSSTTGYY36 pKa = 9.56KK37 pKa = 9.37QASTSFSSLVLPSSSGNYY55 pKa = 9.32NSGGVEE61 pKa = 3.84TTVRR65 pKa = 11.84YY66 pKa = 9.69AISQQGTVSATTSSTDD82 pKa = 4.25LAIQGNGFFVVQSAAGQTFLTRR104 pKa = 11.84SGDD107 pKa = 3.69FSADD111 pKa = 3.01ASGNLVNAAGFTLMGYY127 pKa = 9.77SSASGAPSVVVNGFSGMVPINVTPSGTSVAATTVGSMTGNLNSNATIATSSSTGYY182 pKa = 10.52LPSEE186 pKa = 3.74NTYY189 pKa = 10.67PVTTDD194 pKa = 3.14TSKK197 pKa = 11.38SSITGYY203 pKa = 10.54DD204 pKa = 3.3SQGNAVTFDD213 pKa = 2.93IYY215 pKa = 9.5YY216 pKa = 9.75TKK218 pKa = 10.29TADD221 pKa = 3.66NTWDD225 pKa = 3.44VQVYY229 pKa = 9.27NQADD233 pKa = 4.11LVASPTSGGPLYY245 pKa = 10.94SSAAVGSTEE254 pKa = 3.88MTFDD258 pKa = 4.49DD259 pKa = 5.58DD260 pKa = 4.04GALVSGGTFDD270 pKa = 5.77LSFTSGSTTQSIAMDD285 pKa = 3.5MTGFTQVATDD295 pKa = 4.39FSATGKK301 pKa = 9.59MDD303 pKa = 3.52GQAPNPVTSVTIAKK317 pKa = 9.85DD318 pKa = 3.51GTVSAVYY325 pKa = 10.02KK326 pKa = 10.71DD327 pKa = 3.87SSTKK331 pKa = 9.33EE332 pKa = 3.86LYY334 pKa = 10.14RR335 pKa = 11.84IPLATVASPDD345 pKa = 3.62SLTLEE350 pKa = 4.27SGNVYY355 pKa = 10.22SANGDD360 pKa = 3.45SGVTVTGWPQSGGLGYY376 pKa = 9.71IQSGALEE383 pKa = 4.23EE384 pKa = 4.93SNVDD388 pKa = 4.04LATEE392 pKa = 4.61LTNMITAQKK401 pKa = 10.63SYY403 pKa = 9.31TANSKK408 pKa = 9.73VFQAGSDD415 pKa = 3.75LLDD418 pKa = 3.26VLVNLQRR425 pKa = 4.57
MM1 pKa = 7.61SIFGSMKK8 pKa = 9.46TAISGMNAQANRR20 pKa = 11.84LATVSDD26 pKa = 4.49NIANSSTTGYY36 pKa = 9.56KK37 pKa = 9.37QASTSFSSLVLPSSSGNYY55 pKa = 9.32NSGGVEE61 pKa = 3.84TTVRR65 pKa = 11.84YY66 pKa = 9.69AISQQGTVSATTSSTDD82 pKa = 4.25LAIQGNGFFVVQSAAGQTFLTRR104 pKa = 11.84SGDD107 pKa = 3.69FSADD111 pKa = 3.01ASGNLVNAAGFTLMGYY127 pKa = 9.77SSASGAPSVVVNGFSGMVPINVTPSGTSVAATTVGSMTGNLNSNATIATSSSTGYY182 pKa = 10.52LPSEE186 pKa = 3.74NTYY189 pKa = 10.67PVTTDD194 pKa = 3.14TSKK197 pKa = 11.38SSITGYY203 pKa = 10.54DD204 pKa = 3.3SQGNAVTFDD213 pKa = 2.93IYY215 pKa = 9.5YY216 pKa = 9.75TKK218 pKa = 10.29TADD221 pKa = 3.66NTWDD225 pKa = 3.44VQVYY229 pKa = 9.27NQADD233 pKa = 4.11LVASPTSGGPLYY245 pKa = 10.94SSAAVGSTEE254 pKa = 3.88MTFDD258 pKa = 4.49DD259 pKa = 5.58DD260 pKa = 4.04GALVSGGTFDD270 pKa = 5.77LSFTSGSTTQSIAMDD285 pKa = 3.5MTGFTQVATDD295 pKa = 4.39FSATGKK301 pKa = 9.59MDD303 pKa = 3.52GQAPNPVTSVTIAKK317 pKa = 9.85DD318 pKa = 3.51GTVSAVYY325 pKa = 10.02KK326 pKa = 10.71DD327 pKa = 3.87SSTKK331 pKa = 9.33EE332 pKa = 3.86LYY334 pKa = 10.14RR335 pKa = 11.84IPLATVASPDD345 pKa = 3.62SLTLEE350 pKa = 4.27SGNVYY355 pKa = 10.22SANGDD360 pKa = 3.45SGVTVTGWPQSGGLGYY376 pKa = 9.71IQSGALEE383 pKa = 4.23EE384 pKa = 4.93SNVDD388 pKa = 4.04LATEE392 pKa = 4.61LTNMITAQKK401 pKa = 10.63SYY403 pKa = 9.31TANSKK408 pKa = 9.73VFQAGSDD415 pKa = 3.75LLDD418 pKa = 3.26VLVNLQRR425 pKa = 4.57
Molecular weight: 43.37 kDa
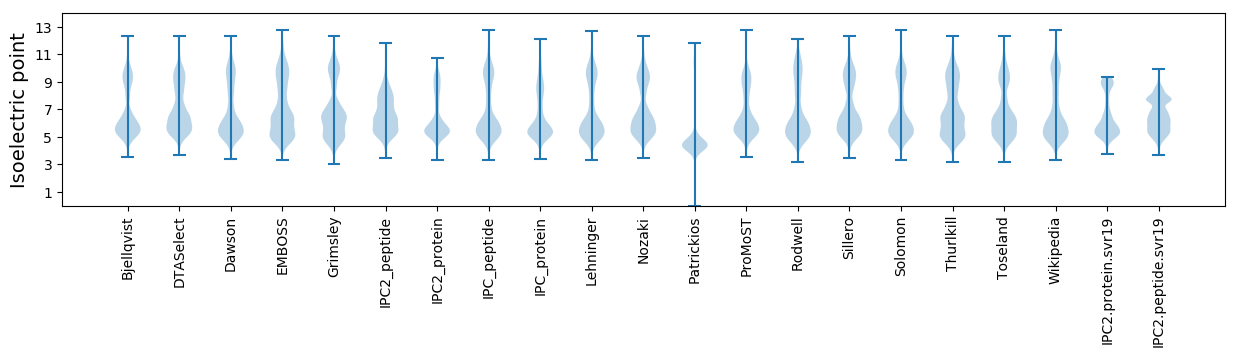
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B9JP40|B9JP40_AGRRK Chaperone protein ClpB OS=Agrobacterium radiobacter (strain K84 / ATCC BAA-868) OX=311403 GN=clpB PE=3 SV=1
MM1 pKa = 7.61SEE3 pKa = 3.66TMLAVLSNIRR13 pKa = 11.84TIDD16 pKa = 3.52DD17 pKa = 3.23MVAAFRR23 pKa = 11.84EE24 pKa = 4.47EE25 pKa = 4.07EE26 pKa = 3.84HH27 pKa = 6.68CRR29 pKa = 11.84RR30 pKa = 11.84LLEE33 pKa = 4.26SMVWRR38 pKa = 11.84DD39 pKa = 3.27GRR41 pKa = 11.84VCPACGYY48 pKa = 9.65KK49 pKa = 10.15RR50 pKa = 11.84SIAIAGRR57 pKa = 11.84DD58 pKa = 3.27MGKK61 pKa = 9.66RR62 pKa = 11.84RR63 pKa = 11.84ARR65 pKa = 11.84PGLYY69 pKa = 9.1QCSSGDD75 pKa = 3.38CRR77 pKa = 11.84FQFTVTTHH85 pKa = 6.24TPLHH89 pKa = 5.59ATKK92 pKa = 10.68LPLRR96 pKa = 11.84TWLKK100 pKa = 11.29AMWLLLQSDD109 pKa = 4.25KK110 pKa = 11.35GLSSVRR116 pKa = 11.84LAEE119 pKa = 4.18TLGVSQPTAWRR130 pKa = 11.84IGHH133 pKa = 7.03ALRR136 pKa = 11.84LMVARR141 pKa = 11.84EE142 pKa = 3.87HH143 pKa = 6.3MLDD146 pKa = 3.26GTVEE150 pKa = 3.75VDD152 pKa = 2.93HH153 pKa = 7.06FYY155 pKa = 11.35LGGRR159 pKa = 11.84PGKK162 pKa = 10.08HH163 pKa = 6.68PDD165 pKa = 3.76DD166 pKa = 4.84PPPGRR171 pKa = 11.84GRR173 pKa = 11.84KK174 pKa = 8.68GQAKK178 pKa = 5.64TQKK181 pKa = 8.29TPVMAIVQRR190 pKa = 11.84PADD193 pKa = 3.86VTPGSPAGDD202 pKa = 3.0ARR204 pKa = 11.84AAVVTGLSLRR214 pKa = 11.84AAVGVIAPQVEE225 pKa = 4.36LHH227 pKa = 6.02AHH229 pKa = 6.05LMSDD233 pKa = 3.4EE234 pKa = 4.33ANAFVAIGEE243 pKa = 4.37SFAVHH248 pKa = 5.02EE249 pKa = 4.75TVNHH253 pKa = 5.93SSRR256 pKa = 11.84EE257 pKa = 4.03YY258 pKa = 10.52VRR260 pKa = 11.84NTVHH264 pKa = 6.19VNSAEE269 pKa = 3.98GFNARR274 pKa = 11.84VRR276 pKa = 11.84RR277 pKa = 11.84TIAGVFHH284 pKa = 7.07HH285 pKa = 7.17ISPEE289 pKa = 3.79LADD292 pKa = 4.86LYY294 pKa = 10.42FHH296 pKa = 7.44EE297 pKa = 5.62IGFRR301 pKa = 11.84WSQRR305 pKa = 11.84VVIGQAVRR313 pKa = 11.84RR314 pKa = 11.84NRR316 pKa = 11.84SGKK319 pKa = 10.05EE320 pKa = 3.34SGKK323 pKa = 9.89ILWSRR328 pKa = 11.84VPPALQLLQVFRR340 pKa = 11.84AATGRR345 pKa = 11.84QMRR348 pKa = 11.84RR349 pKa = 11.84SHH351 pKa = 6.14QGGITIKK358 pKa = 9.83STVAVFGG365 pKa = 3.86
MM1 pKa = 7.61SEE3 pKa = 3.66TMLAVLSNIRR13 pKa = 11.84TIDD16 pKa = 3.52DD17 pKa = 3.23MVAAFRR23 pKa = 11.84EE24 pKa = 4.47EE25 pKa = 4.07EE26 pKa = 3.84HH27 pKa = 6.68CRR29 pKa = 11.84RR30 pKa = 11.84LLEE33 pKa = 4.26SMVWRR38 pKa = 11.84DD39 pKa = 3.27GRR41 pKa = 11.84VCPACGYY48 pKa = 9.65KK49 pKa = 10.15RR50 pKa = 11.84SIAIAGRR57 pKa = 11.84DD58 pKa = 3.27MGKK61 pKa = 9.66RR62 pKa = 11.84RR63 pKa = 11.84ARR65 pKa = 11.84PGLYY69 pKa = 9.1QCSSGDD75 pKa = 3.38CRR77 pKa = 11.84FQFTVTTHH85 pKa = 6.24TPLHH89 pKa = 5.59ATKK92 pKa = 10.68LPLRR96 pKa = 11.84TWLKK100 pKa = 11.29AMWLLLQSDD109 pKa = 4.25KK110 pKa = 11.35GLSSVRR116 pKa = 11.84LAEE119 pKa = 4.18TLGVSQPTAWRR130 pKa = 11.84IGHH133 pKa = 7.03ALRR136 pKa = 11.84LMVARR141 pKa = 11.84EE142 pKa = 3.87HH143 pKa = 6.3MLDD146 pKa = 3.26GTVEE150 pKa = 3.75VDD152 pKa = 2.93HH153 pKa = 7.06FYY155 pKa = 11.35LGGRR159 pKa = 11.84PGKK162 pKa = 10.08HH163 pKa = 6.68PDD165 pKa = 3.76DD166 pKa = 4.84PPPGRR171 pKa = 11.84GRR173 pKa = 11.84KK174 pKa = 8.68GQAKK178 pKa = 5.64TQKK181 pKa = 8.29TPVMAIVQRR190 pKa = 11.84PADD193 pKa = 3.86VTPGSPAGDD202 pKa = 3.0ARR204 pKa = 11.84AAVVTGLSLRR214 pKa = 11.84AAVGVIAPQVEE225 pKa = 4.36LHH227 pKa = 6.02AHH229 pKa = 6.05LMSDD233 pKa = 3.4EE234 pKa = 4.33ANAFVAIGEE243 pKa = 4.37SFAVHH248 pKa = 5.02EE249 pKa = 4.75TVNHH253 pKa = 5.93SSRR256 pKa = 11.84EE257 pKa = 4.03YY258 pKa = 10.52VRR260 pKa = 11.84NTVHH264 pKa = 6.19VNSAEE269 pKa = 3.98GFNARR274 pKa = 11.84VRR276 pKa = 11.84RR277 pKa = 11.84TIAGVFHH284 pKa = 7.07HH285 pKa = 7.17ISPEE289 pKa = 3.79LADD292 pKa = 4.86LYY294 pKa = 10.42FHH296 pKa = 7.44EE297 pKa = 5.62IGFRR301 pKa = 11.84WSQRR305 pKa = 11.84VVIGQAVRR313 pKa = 11.84RR314 pKa = 11.84NRR316 pKa = 11.84SGKK319 pKa = 10.05EE320 pKa = 3.34SGKK323 pKa = 9.89ILWSRR328 pKa = 11.84VPPALQLLQVFRR340 pKa = 11.84AATGRR345 pKa = 11.84QMRR348 pKa = 11.84RR349 pKa = 11.84SHH351 pKa = 6.14QGGITIKK358 pKa = 9.83STVAVFGG365 pKa = 3.86
Molecular weight: 40.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2107362 |
30 |
4272 |
315.8 |
34.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.653 ± 0.043 | 0.785 ± 0.009 |
5.674 ± 0.022 | 5.486 ± 0.032 |
3.93 ± 0.022 | 8.273 ± 0.032 |
2.058 ± 0.014 | 5.929 ± 0.022 |
3.718 ± 0.025 | 10.051 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.521 ± 0.015 | 2.933 ± 0.023 |
4.861 ± 0.02 | 3.2 ± 0.021 |
6.513 ± 0.036 | 5.993 ± 0.024 |
5.477 ± 0.026 | 7.29 ± 0.021 |
1.291 ± 0.012 | 2.363 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
